













| General Information: |
|
| Name(s) found: |
LRKS7_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 681 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endomembrane system
[IEA]
|
| Biological Process: |
protein amino acid phosphorylation
[IEA]
|
| Molecular Function: |
kinase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLSRKLLVI FFTWITALSM SKPIFVSSDN MNFTFKSFTI RNLTFLGDSH LRNGVVGLTR 60
61 ELGVPDTSSG TVIYNNPIRF YDPDSNTTAS FSTHFSFTVQ NLNPDPTSAG DGLAFFLSHD 120
121 NDTLGSPGGY LGLVNSSQPM KNRFVAIEFD TKLDPHFNDP NGNHIGLDVD SLNSISTSDP 180
181 LLSSQIDLKS GKSITSWIDY KNDLRLLNVF LSYTDPVTTT KKPEKPLLSV NIDLSPFLNG 240
241 EMYVGFSGST EGSTEIHLIE NWSFKTSGFL PVRSKSNHLH NVSDSSVVND DPVVIPSKKR 300
301 RHRHNLAIGL GISCPVLICL ALFVFGYFTL KKWKSVKAEK ELKTELITGL REFSYKELYT 360
361 ATKGFHSSRV IGRGAFGNVY RAMFVSSGTI SAVKRSRHNS TEGKTEFLAE LSIIACLRHK 420
421 NLVQLQGWCN EKGELLLVYE FMPNGSLDKI LYQESQTGAV ALDWSHRLNI AIGLASALSY 480
481 LHHECEQQVV HRDIKTSNIM LDINFNARLG DFGLARLTEH DKSPVSTLTA GTMGYLAPEY 540
541 LQYGTATEKT DAFSYGVVIL EVACGRRPID KEPESQKTVN LVDWVWRLHS EGRVLEAVDE 600
601 RLKGEFDEEM MKKLLLVGLK CAHPDSNERP SMRRVLQILN NEIEPSPVPK MKPTLSFSCG 660
661 LSLDDIVSED EEGDSIVYVV S |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
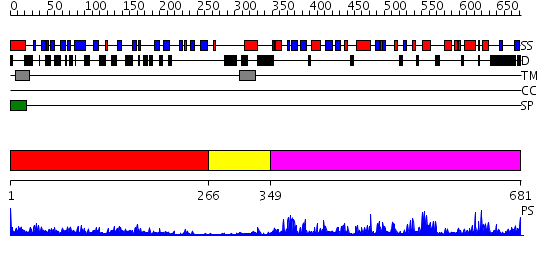
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..265] | 31.0 | Phytohemagglutinin-L, PHA-L, also arcelin | |
| 2 | View Details | [266..348] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [349..681] | 70.221849 | No description for 2qkwB was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|