













| General Information: |
|
| Name(s) found: |
PUB12_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 654 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ubiquitin ligase complex
[IEA]
ribosome [IEA] intracellular [IEA] |
| Biological Process: |
response to chitin
[IEP]
|
| Molecular Function: |
ubiquitin-protein ligase activity
[IEA]
structural constituent of ribosome [IEA] binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKSEKHKLA QTLIDSINEI ASISDSVTPM KKHCANLSRR LSLLLPMLEE IRDNQESSSE 60
61 VVNALLSVKQ SLLHAKDLLS FVSHVSKIYL VLERDQVMVK FQKVTSLLEQ ALSIIPYENL 120
121 EISDELKEQV ELVLVQLRRS LGKRGGDVYD DELYKDVLSL YSGRGSVMES DMVRRVAEKL 180
181 QLMTITDLTQ ESLALLDMVS SSGGDDPGES FEKMSMVLKK IKDFVQTYNP NLDDAPLRLK 240
241 SSLPKSRDDD RDMLIPPEEF RCPISLELMT DPVIVSSGQT YERECIKKWL EGGHLTCPKT 300
301 QETLTSDIMT PNYVLRSLIA QWCESNGIEP PKRPNISQPS SKASSSSSAP DDEHNKIEEL 360
361 LLKLTSQQPE DRRSAAGEIR LLAKQNNHNR VAIAASGAIP LLVNLLTISN DSRTQEHAVT 420
421 SILNLSICQE NKGKIVYSSG AVPGIVHVLQ KGSMEARENA AATLFSLSVI DENKVTIGAA 480
481 GAIPPLVTLL SEGSQRGKKD AATALFNLCI FQGNKGKAVR AGLVPVLMRL LTEPESGMVD 540
541 ESLSILAILS SHPDGKSEVG AADAVPVLVD FIRSGSPRNK ENSAAVLVHL CSWNQQHLIE 600
601 AQKLGIMDLL IEMAENGTDR GKRKAAQLLN RFSRFNDQQK QHSGLGLEDQ ISLI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
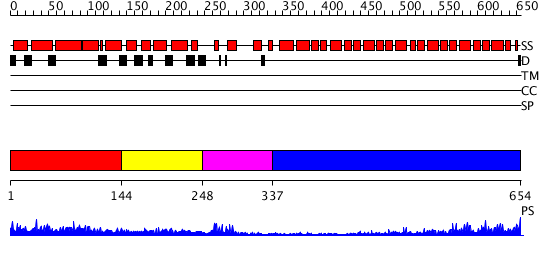
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..143] | 3.112995 | View MSA. Confident ab initio structure predictions are available. | |
| 2 | View Details | [144..247] | 1.185996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [248..336] | 9.221849 | NMR solution structure of the U box domain from AtPUB14, an armadillo repeat containing protein from Arabidopsis thaliana | |
| 4 | View Details | [337..654] | 33.522879 | beta-Catenin |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)