













| General Information: |
|
| Name(s) found: |
gi|15219728
[NCBI NR]
gi|12597768 [NCBI NR] |
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1417 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
|
| Biological Process: |
DNA repair
[IEA][ISS]
DNA replication [IEA][ISS] DNA recombination [IEA][ISS] |
| Molecular Function: |
ATP binding
[IEA][ISS]
DNA binding [IEA] DNA ligase (ATP) activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASDSAGATI SGNFSNSDNS ETLNLNTTKL YSSAISSISP QFPSPKPTSS CPSIPNSKRI 60
61 PNTNFIVDLF RLPHQSSSVA FFLSHFHSDH YSGLSSSWSK GIIYCSHKTA RLVAEILQVP 120
121 SQFVFALPMN QMVKIDGSEV VLIEANHCPG AVQFLFKVKL ESSGFEKYVH TGDFRFCDEM 180
181 RFDPFLNGFV GCDGVFLDTT YCNPKFVFPS QEESVGYVVS VIDKISEEKV LFLVATYVVG 240
241 KEKILVEIAR RCKRKIVVDA RKMSMLSVLG CGEEGMFTED ENESDVHVVG WNVLGETWPY 300
301 FRPNFVKMNE IMVEKGYDKV VGFVPTGWTY EVKRNKFAVR FKDSMEIHLV PYSEHSNYDE 360
361 LREFIKFLKP KRVIPTVGVD IEKFDCKEVN KMQKHFSGLV DEMANKKDFL LGFYRQSYQK 420
421 NEKSDVDVVS HSAEVYEEEE KNACEDGGEN VPSSRGPILH DTTPSSDSRL LIKLRDSLPA 480
481 WVTEEQMLDL IKKHAGNPVD IVSNFYEYEA ELYKQASLPT PSLNNQAVLF DDDVTDLQPN 540
541 PVKGICPDVQ AIQKGFDLPR KMNLTKGTIS PGKRGKSSGS KSNKKAKKDP KSKPVGPGQP 600
601 TLFKFFNKVL DGGSNSVSVG SETEECNTDK KMVHIDASEA YKEVTDQFID IVNGSESLRD 660
661 YAASIIDEAK GDISRALNIY YSKPREIPGD HAGERGLSSK TIQYPKCSEA CSSQEDKKAS 720
721 ENSGHAVNIC VQTSAEESVD KNYVSLPPEK YQPKEHACWR EGQPAPYIHL VRTFASVESE 780
781 KGKIKAMSML CNMFRSLFAL SPEDVLPAVY LCTNKIAADH ENIELNIGGS LISSALEEAC 840
841 GISRSTVRDM YNSLGDLGDV AQLCRQTQKL LVPPPPLLVR DVFSTLRKIS VQTGTGSTRL 900
901 KKNLIVKLMR SCREKEIKFL VRTLARNLRI GAMLRTVLPA LGRAIVMNSF WNDHNKELSE 960
961 SCFREKLEGV SAAVVEAYNI LPSLDVVVPS LMDKDIEFST STLSMVPGIP IKPMLAKIAK1020
1021 GVQEFFNLSQ EKAFTCEYKY DGQRAQIHKL LDGTVCIFSR NGDETTSRFP DLVDVIKQFS1080
1081 CPAAETFMLD AEVVATDRIN GNKLMSFQEL STRERGSKDA LITTESIKVE VCVFVFDIMF1140
1141 VNGEQLLALP LRERRRRLKE VFPETRPGYL EYAKEITVSA TVGAEEASLN NHDTLSRINA1200
1201 FLEEAFQSSC EGIMVKSLDV NAGYCPTKRS DSWLKVKRDY VDGLGDTLDL VPIGAWYGNG1260
1261 RKAGWYSPFL MACFNPETEE FQSVCRVMSG FSDAFYIEVM KSSTLTLLFN DETDGMKEFY1320
1321 SEDKILAKKP PYYRTGETPD MWFSAEVVWE IRGADFTVSP VHSASLGLVH PSRGISVRFP1380
1381 RFISKVTDRN PEECSTATDI AEMFHAQTRK MNITSQH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
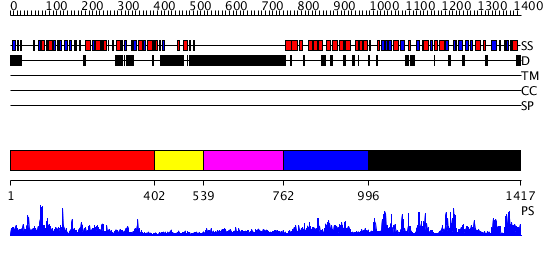
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..401] | 11.221849 | Rubredoxin oxygen:oxidoreductase (ROO), C-terminal domain; Rubredoxin oxygen:oxidoreductase (ROO), N-terminal domain | |
| 2 | View Details | [402..538] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [539..761] | 3.139995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [762..995] | 3.221849 | The crystal structure of murine p97/VCP at 3.6A | |
| 5 | View Details | [996..1417] | 33.522879 | ATP-DEPENDENT DNA LIGASE FROM BACTERIOPHAGE T7 COMPLEX WITH ATP |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)