













| General Information: |
|
| Name(s) found: |
M3KE2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1367 amino acids |
Gene Ontology: |
|
| Cellular Component: |
vacuole
[IDA]
|
| Biological Process: |
plasma membrane organization
[IGI]
pollen development [IGI] |
| Molecular Function: |
ATP binding
[IEA]
protein tyrosine kinase activity [IEA] protein kinase activity [IEA] binding [IEA] protein serine/threonine kinase activity [IEA] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MARQMTSSQF HKSKTLDNKY MLGDEIGKGA YGRVYIGLDL ENGDFVAIKQ VSLENIGQED 60
61 LNTIMQEIDL LKNLNHKNIV KYLGSLKTKT HLHIILEYVE NGSLANIIKP NKFGPFPESL 120
121 VTVYIAQVLE GLVYLHEQGV IHRDIKGANI LTTKEGLVKL ADFGVATKLN EADFNTHSVV 180
181 GTPYWMAPEV IELSGVCAAS DIWSVGCTII ELLTCVPPYY DLQPMPALYR IVQDDTPPIP 240
241 DSLSPDITDF LRLCFKKDSR QRPDAKTLLS HPWIRNSRRA LRSSLRHSGT IRYMKETDSS 300
301 SEKDAEGSQE VVESVSAEKV EVTKTNSKSK LPVIGGASFR SEKDQSSPSD LGEEGTDSED 360
361 DINSDQGPTL SMHDKSSRQS GTCSISSDAK GTSQDVLENH EKYDRDEIPG NLETEASEGR 420
421 RNTLATKLVG KEYSIQSSHS FSQKGEDGLR KAVKTPSSFG GNELTRFSDP PGDASLHDLF 480
481 HPLDKVPEGK TNEASTSTPT ANVNQGDSPV ADGGKNDLAT KLRARIAQKQ MEGETGHSQD 540
541 GGDLFRLMMG VLKDDVLNID DLVFDEKVPP ENLFPLQAVE FSRLVSSLRP DESEDAIVTS 600
601 SLKLVAMFRQ RPGQKAVFVT QNGFLPLMDL LDIPKSRVIC AVLQLINEIV KDNTDFLENA 660
661 CLVGLIPLVM SFAGFERDRS REIRKEAAYF LQQLCQSSPL TLQMFISCRG IPVLVGFLEA 720
721 DYAKHREMVH LAIDGMWQVF KLKKSTSRND FCRIAAKNGI LLRLVNTLYS LSEATRLASI 780
781 SGDALILDGQ TPRARSGQLD PNNPIFSQRE TSPSVIDHPD GLKTRNGGGE EPSHALTSNS 840
841 QSSDVHQPDA LHPDGDRPRL SSVVADATED VIQQHRISLS ANRTSTDKLQ KLAEGASNGF 900
901 PVTQPDQVRP LLSLLEKEPP SRKISGQLDY VKHIAGIERH ESRLPLLYAS DEKKTNGDLE 960
961 FIMAEFAEVS GRGKENGNLD TAPRYSSKTM TKKVMAIERV ASTCGIASQT ASGVLSGSGV1020
1021 LNARPGSTTS SGLLAHALSA DVSMDYLEKV ADLLLEFARA ETTVKSYMCS QSLLSRLFQM1080
1081 FNRVEPPILL KILECTNHLS TDPNCLENLQ RADAIKQLIP NLELKEGPLV YQIHHEVLSA1140
1141 LFNLCKINKR RQEQAAENGI IPHLMLFVMS DSPLKQYALP LLCDMAHASR NSREQLRAHG1200
1201 GLDVYLSLLD DEYWSVIALD SIAVCLAQDV DQKVEQAFLK KDAIQKLVNF FQNCPERHFV1260
1261 HILEPFLKII TKSSSINKTL ALNGLTPLLI ARLDHQDAIA RLNLLKLIKA VYEKHPKPKQ1320
1321 LIVENDLPQK LQNLIEERRD GQRSGGQVLV KQMATSLLKA LHINTIL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
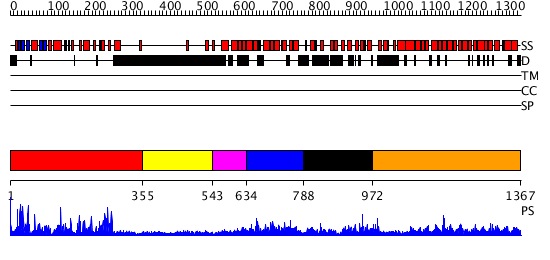
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..354] | 74.522879 | No description for 3comA was found. | |
| 2 | View Details | [355..542] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [543..633] | 9.69897 | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3 | |
| 4 | View Details | [634..787] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [788..971] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [972..1367] | 73.0 | No description for 2jdqA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)