













| General Information: |
|
| Name(s) found: |
DGP10_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 586 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
proteolysis
[IEA][ISS]
|
| Molecular Function: |
protein binding
[IEA]
catalytic activity [IEA] serine-type endopeptidase activity [IEA] serine-type peptidase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLLRSFRTVE LLRRISTSSV SGYRTSPSLL QRCNGFQSYL PHRVTTTESP FPSHISRFCS 60
61 SQSANSQNEN RHTTLSSPVS SRRVNNRKIS RRRKAGKSLS ISPAADAVDL ALDSVVKIFT 120
121 VSTSPSYFLP WQNKSQRESM GSGFVISGRK IITNAHVVAD HSFVLVRKHG SSIKHRAEVQ 180
181 AVGHECDLAI LVVDSEVFWE GMNALELGDI PFLQEAVAVV GYPQGGDNIS VTKGVVSRVE 240
241 PTQYVHGATQ LMAIQIDAAI NPGNSGGPAI MGNKVAGVAF QNLSGAENIG YIIPTPVIKH 300
301 FINGVEECGK YIGFCSMGVS CQPMENGELR SGFQMSSEMT GVLVSKINPL SDAHKILKKD 360
361 DVLLAFDGVP IANDGTVPFR NRERITFDHL VSMKKPDETA LVKVLREGKE HEFSITLRPL 420
421 QPLVPVHQFD QLPSYYIFAG FVFVPLTQPY LHEYGEDWYN TSPRTLCHRA LKDLPKKAGQ 480
481 QLVIVSQVLM DDINTGYERL AELQVNKVNG VEVNNLRHLC QLIENCNTEK LRIDLDDESR 540
541 VIVLNYQSAK IATSLILKRH RIASAISSDL LIEQNLETEL ASCSAV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
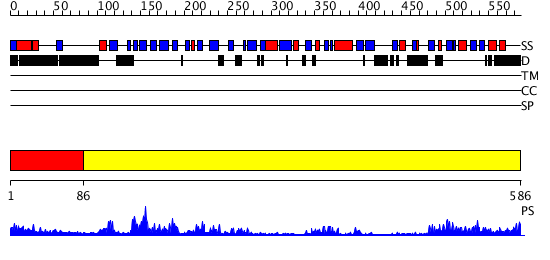
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..85] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [86..586] | 64.30103 | Protease Do (DegP, HtrA), C-terminal domains; Protease Do (DegP, HtrA), catalytic domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)