













| General Information: |
|
| Name(s) found: |
CALSC_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1780 amino acids |
Gene Ontology: |
|
| Cellular Component: |
1,3-beta-glucan synthase complex
[ISS]
plasma membrane [IDA] |
| Biological Process: |
1,3-beta-glucan biosynthetic process
[ISS]
defense response signaling pathway, resistance gene-dependent [IMP] leaf senescence [IMP] salicylic acid mediated signaling pathway [IEP] pollen development [IGI] response to fungus [IMP] defense response [IMP] defense response to bacterium [IMP] leaf morphogenesis [IMP] reproduction [IGI] callose deposition in cell wall during defense response [IMP] callose deposition during defense response [IMP] [NOT]defense response to fungus [IMP] |
| Molecular Function: |
1,3-beta-glucan synthase activity
[ISS]
transferase activity, transferring glycosyl groups [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLRHRTVPP QTGRPLAAEA VGIEEEPYNI IPVNNLLADH PSLRFPEVRA AAAALKTVGD 60
61 LRRPPYVQWR SHYDLLDWLA LFFGFQKDNV RNQREHMVLH LANAQMRLSP PPDNIDSLDS 120
121 AVVRRFRRKL LANYSSWCSY LGKKSNIWIS DRNPDSRREL LYVGLYLLIW GEAANLRFMP 180
181 ECICYIFHNM ASELNKILED CLDENTGQPY LPSLSGENAF LTGVVKPIYD TIQAEIDESK 240
241 NGTVAHCKWR NYDDINEYFW TDRCFSKLKW PLDLGSNFFK SRGKSVGKTG FVERRTFFYL 300
301 YRSFDRLWVM LALFLQAAII VAWEEKPDTS SVTRQLWNAL KARDVQVRLL TVFLTWSGMR 360
361 LLQAVLDAAS QYPLVSRETK RHFFRMLMKV IAAAVWIVAF TVLYTNIWKQ KRQDRQWSNA 420
421 ATTKIYQFLY AVGAFLVPEI LALALFIIPW MRNFLEETNW KIFFALTWWF QGKSFVGRGL 480
481 REGLVDNIKY STFWIFVLAT KFTFSYFLQV KPMIKPSKLL WNLKDVDYEW HQFYGDSNRF 540
541 SVALLWLPVV LIYLMDIQIW YAIYSSIVGA VVGLFDHLGE IRDMGQLRLR FQFFASAIQF 600
601 NLMPEEQLLN ARGFGNKFKD GIHRLKLRYG FGRPFKKLES NQVEANKFAL IWNEIILAFR 660
661 EEDIVSDREV ELLELPKNSW DVTVIRWPCF LLCNELLLAL SQARELIDAP DKWLWHKICK 720
721 NEYRRCAVVE AYDSIKHLLL SIIKVDTEEH SIITVFFQII NQSIQSEQFT KTFRVDLLPK 780
781 IYETLQKLVG LVNDEETDSG RVVNVLQSLY EIATRQFFIE KKTTEQLSNE GLTPRDPASK 840
841 LLFQNAIRLP DASNEDFYRQ VRRLHTILTS RDSMHSVPVN LEARRRIAFF SNSLFMNMPH 900
901 APQVEKMMAF SVLTPYYSEE VVYSKEQLRN ETEDGISTLY YLQTIYADEW KNFKERMHRE 960
961 GIKTDSELWT TKLRDLRLWA SYRGQTLART VRGMMYYYRA LKMLAFLDSA SEMDIREGAQ1020
1021 ELGSVRNLQG ELGGQSDGFV SENDRSSLSR ASSSVSTLYK GHEYGTALMK FTYVVACQIY1080
1081 GSQKAKKEPQ AEEILYLMKQ NEALRIAYVD EVPAGRGETD YYSVLVKYDH QLEKEVEIFR1140
1141 VKLPGPVKLG EGKPENQNHA MIFTRGDAVQ TIDMNQDSYF EEALKMRNLL QEYNHYHGIR1200
1201 KPTILGVREH IFTGSVSSLA WFMSAQETSF VTLGQRVLAN PLKVRMHYGH PDVFDRFWFL1260
1261 SRGGISKASR VINISEDIFA GFNCTLRGGN VTHHEYIQVG KGRDVGLNQI SMFEAKVASG1320
1321 NGEQVLSRDV YRLGHRLDFF RMLSFFYTTV GFFFNTMMVI LTVYAFLWGR VYLALSGVEK1380
1381 SALADSTDTN AALGVILNQQ FIIQLGLFTA LPMIVEWSLE EGFLLAIWNF IRMQIQLSAV1440
1441 FYTFSMGTRA HYFGRTILHG GAKYRATGRG FVVEHKGFTE NYRLYARSHF VKAIELGLIL1500
1501 IVYASHSPIA KDSLIYIAMT ITSWFLVISW IMAPFVFNPS GFDWLKTVYD FEDFMNWIWY1560
1561 QGRISTKSEQ SWEKWWYEEQ DHLRNTGKAG LFVEIILVLR FFFFQYGIVY QLKIANGSTS1620
1621 LFVYLFSWIY IFAIFVLFLV IQYARDKYSA KAHIRYRLVQ FLLIVLAILV IVALLEFTHF1680
1681 SFIDIFTSLL AFIPTGWGIL LIAQTQRKWL KNYTIFWNAV VSVARMYDIL FGILIMVPVA1740
1741 FLSWMPGFQS MQTRILFNEA FSRGLRIMQI VTGKKSKGDV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
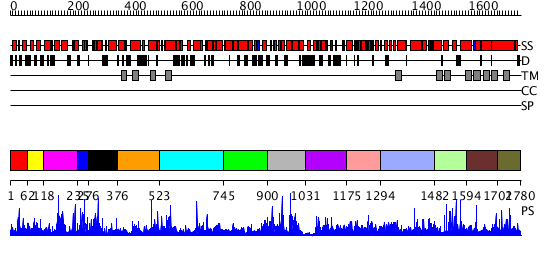
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..61] | 1.016996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [62..117] | 2.023984 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [118..234] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [235..275] | 1.026989 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [276..375] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [376..522] | 1.024986 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [523..744] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [745..899] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [900..1030] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1031..1174] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1175..1293] | 3.053959 | View MSA. No confident structure predictions are available. | |
| 12 | View Details | [1294..1481] | 2.055944 | View MSA. No confident structure predictions are available. | |
| 13 | View Details | [1482..1593] | N/A | No confident structure predictions are available. | |
| 14 | View Details | [1594..1701] | 2.045971 | View MSA. No confident structure predictions are available. | |
| 15 | View Details | [1702..1780] | 1.040982 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|