













| General Information: |
|
| Name(s) found: |
DNJ19_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 346 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum lumen
[IDA]
plasma membrane [IDA] |
| Biological Process: |
protein folding
[ISS]
|
| Molecular Function: |
heat shock protein binding
[IEA]
unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAIRWSELCI VLFALSYAIC VLAGKSYYDV LQVPKGASDE QIKRAYRKLA LKYHPDKNQG 60
61 NEEATRKFAE INNAYEVLSD EEKREIYNKY GEEGLKQFSA NGGRGGGGGG MNMQDIFSSF 120
121 FGGGSMEEEE KVVKGDDVIV ELEATLEDLY MGGSMKVWRE KNVIKPAPGK RKCNCRNEVY 180
181 HRQIGPGMFQ QMTEQVCDKC PNVKYEREGY FVTVDIEKGM KDGEEVSFYE DGEPILDGDP 240
241 GDLKFRIRTA PHARFRRDGN DLHMNVNITL VEALVGFEKS FKHLDDHEVD ISSKGITKPK 300
301 EVKKFKGEGM PLHYSTKKGN LFVTFEVLFP SSLTDDQKKK IKEVFA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
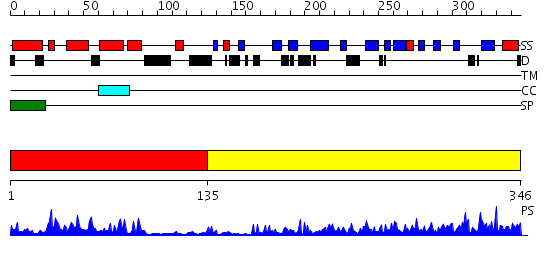
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..134] | 25.522879 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 2 | View Details | [135..346] | 44.221849 | The crystal structure of Hsp40 Ydj1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|