













| General Information: |
|
| Name(s) found: |
SCC13_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 693 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
nuclear cohesin complex [ISS] nucleolus [IDA] |
| Biological Process: |
megagametogenesis
[IMP]
pollen development [IMP] mitosis [IEP] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFYSHTLLAR KGPLGTVWCA AHVHQRLKKS QYTSINIPDT VDNIMFPEVP LALRTSSHLL 60
61 VGVVRIYSKK VDYLYNDWNL LNTWVAKAFV STQVNLPEDA RQAPPESVTL PQALNLDEFD 120
121 LEDDTLDMEF DNHTRSEEDI TLTDQIPTGI DPYVAVTFDE DIISESIPMD VDQSTEPVSR 180
181 HTGEIDVETA HETGPDNEPR DSNIAFDTGT YSPRNVTEEF TEVQDPRQSN LTEERIPNSE 240
241 RNDATSPGTV PEIERMRDAA HDLSPTSHPS FAAQQQDVRV ERTESLDETL NEKEPTIPSI 300
301 DEEMLNSGRH SAFELRSGSP GSAAGSEEER ADFVHPSPQL VLQPSPPPQP QRRARKRKNF 360
361 DGVTVLTNKN ISERLKDPSD TLRKRKKMPS SKLKFWRMNN QSRKDQNFNE PLFTGFSDDL 420
421 RNVFEKDYVA SKPHLAVSDE TLPEPASVSP TREAEVEINP VSPIPDSTNP DSTVQLSPAQ 480
481 QTEDVLDSAG PRPAHAESVA TEAQSPRTFD NDDMGIEHLR DGGFPVYMPS PPPRSSPFRT 540
541 DDFTTQSGNW ETESYRTEPS TSTVPEDLPG QRNLGLSPVS ERTDEELYFL EVGGNSPVGT 600
601 PASQDSAALT GRARALAQYL KQRSSSSPTT SSHPSGDLSL SEILAGKTRK LAARMFFETL 660
661 VLKSRGLIDM QQDRPYGDIA LKLMPALFSK VQT |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
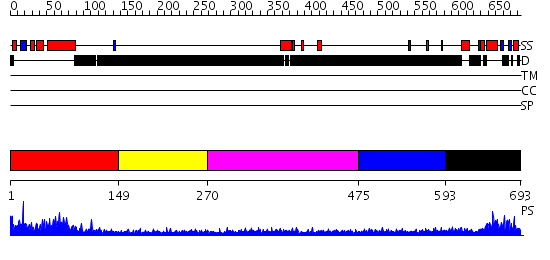
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..148] | 65.619789 | No description for PF04825.4 was found. No confident structure predictions are available. | |
| 2 | View Details | [149..269] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [270..474] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [475..592] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [593..693] | 15.39794 | Sc Smc1hd:Scc1-C complex, ATPgS |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. | |||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)