













| General Information: |
|
| Name(s) found: |
R13L4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 852 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
apoptosis
[IEA]
defense response [ISS] |
| Molecular Function: |
ATP binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVDAVVTVFL EKTLNILEEK GRTVSDYRKQ LEDLQSELKY MQSFLKDAER QKRTNETLRT 60
61 LVADLRELVY EAEDILVDCQ LADGDDGNEQ RSSNAWLSRL HPARVPLQYK KSKRLQEINE 120
121 RITKIKSQVE PYFEFITPSN VGRDNGTDRW SSPVYDHTQV VGLEGDKRKI KEWLFRSNDS 180
181 QLLIMAFVGM GGLGKTTIAQ EVFNDKEIEH RFERRIWVSV SQTFTEEQIM RSILRNLGDA 240
241 SVGDDIGTLL RKIQQYLLGK RYLIVMDDVW DKNLSWWDKI YQGLPRGQGG SVIVTTRSES 300
301 VAKRVQARDD KTHRPELLSP DNSWLLFCNV AFAANDGTCE RPELEDVGKE IVTKCKGLPL 360
361 TIKAVGGLLL CKDHVYHEWR RIAEHFQDEL RGNTSETDNV MSSLQLSYDE LPSHLKSCIL 420
421 TLSLYPEDCV IPKQQLVHGW IGEGFVMWRN GRSATESGED CFSGLTNRCL IEVVDKTYSG 480
481 TIITCKIHDM VRDLVIDIAK KDSFSNPEGL NCRHLGISGN FDEKQIKVNH KLRGVVSTTK 540
541 TGEVNKLNSD LAKKFTDCKY LRVLDISKSI FDAPLSEILD EIASLQHLAC LSLSNTHPLI 600
601 QFPRSMEDLH NLQILDASYC QNLKQLQPCI VLFKKLLVLD MTNCGSLECF PKGIGSLVKL 660
661 EVLLGFKPAR SNNGCKLSEV KNLTNLRKLG LSLTRGDQIE EEELDSLINL SKLMSISINC 720
721 YDSYGDDLIT KIDALTPPHQ LHELSLQFYP GKSSPSWLSP HKLPMLRYMS ICSGNLVKMQ 780
781 EPFWGNENTH WRIEGLMLSS LSDLDMDWEV LQQSMPYLRT VTANWCPELE SFAIEDVGFR 840
841 GGVWMKTPLH RT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
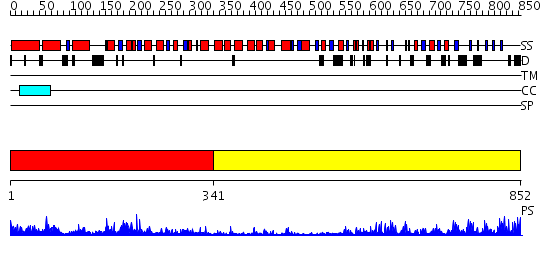
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..340] | 4.0 | Crystal Structure Analysis of ClpB | |
| 2 | View Details | [341..852] | 5.39794 | Rab geranylgeranyltransferase alpha-subunit, N-terminal domain; Rab geranylgeranyltransferase alpha-subunit, insert domain; Rab geranylgeranyltransferase alpha-subunit, C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)