













| General Information: |
|
| Name(s) found: |
PRL1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 486 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
CUL4 RING ubiquitin ligase complex [IPI] |
| Biological Process: |
fruit development
[IMP]
defense response signaling pathway, resistance gene-dependent [IMP] sugar mediated signaling pathway [IMP] defense response to bacterium [IMP] cotyledon development [IMP] hormone-mediated signaling pathway [IMP] root development [IMP] response to glucose stimulus [IMP] negative regulation of transcription [IMP] leaf development [IMP] proteolysis [IMP] defense response signaling pathway, resistance gene-independent [IMP] defense response to fungus [IMP] |
| Molecular Function: |
nucleotide binding
[ISS]
protein binding [IPI] basal transcription repressor activity [IMP][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPAPTTEIEP IEAQSLKKLS LKSLKRSLEL FSPVHGQFPP PDPEAKQIRL SHKMKVAFGG 60
61 VEPVVSQPPR QPDRINEQPG PSNALSLAAP EGSKSTQKGA TESAIVVGPT LLRPILPKGL 120
121 NYTGSSGKST TIIPANVSSY QRNLSTAALM ERIPSRWPRP EWHAPWKNYR VIQGHLGWVR 180
181 SVAFDPSNEW FCTGSADRTI KIWDVATGVL KLTLTGHIEQ VRGLAVSNRH TYMFSAGDDK 240
241 QVKCWDLEQN KVIRSYHGHL SGVYCLALHP TLDVLLTGGR DSVCRVWDIR TKMQIFALSG 300
301 HDNTVCSVFT RPTDPQVVTG SHDTTIKFWD LRYGKTMSTL THHKKSVRAM TLHPKENAFA 360
361 SASADNTKKF SLPKGEFCHN MLSQQKTIIN AMAVNEDGVM VTGGDNGSIW FWDWKSGHSF 420
421 QQSETIVQPG SLESEAGIYA ACYDNTGSRL VTCEADKTIK MWKEDENATP ETHPINFKPP 480
481 KEIRRF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
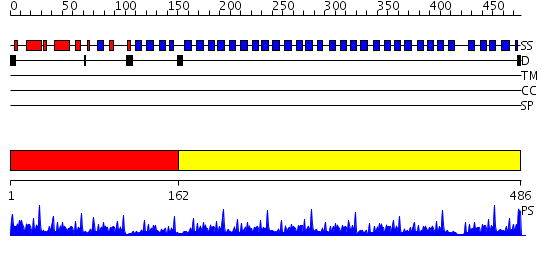
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..161] | 28.045757 | No description for 2pm9A was found. | |
| 2 | View Details | [162..486] | 82.69897 | No description for 2gnqA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)