













| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVSGVGGSGG GRGGGRGGEE EPSSSHTPNN RRGGEQAQSS GTKSLRPRSN TESMSKAIQQ 60
61 YTVDARLHAV FEQSGESGKS FDYSQSLKTT TYGSSVPEQQ ITAYLSRIQR GGYIQPFGCM 120
121 IAVDESSFRI IGYSENAREM LGIMPQSVPT LEKPEILAMG TDVRSLFTSS SSILLERAFV 180
181 AREITLLNPV WIHSKNTGKP FYAILHRIDV GVVIDLEPAR TEDPALSIAG AVQSQKLAVR 240
241 AISQLQALPG GDIKLLCDTV VESVRDLTGY DRVMVYKFHE DEHGEVVAES KRDDLEPYIG 300
301 LHYPATDIPQ ASRFLFKQNR VRMIVDCNAT PVLVVQDDRL TQSMCLVGST LRAPHGCHSQ 360
361 YMANMGSIAS LAMAVIINGN EDDGSNVASG RSSMRLWGLV VCHHTSSRCI PFPLRYACEF 420
421 LMQAFGLQLN MELQLALQMS EKRVLRTQTL LCDMLLRDSP AGIVTQSPSI MDLVKCDGAA 480
481 FLYHGKYYPL GVAPSEVQIK DVVEWLLANH ADSTGLSTDS LGDAGYPGAA ALGDAVCGMA 540
541 VAYITKRDFL FWFRSHTAKE IKWGGAKHHP EDKDDGQRMH PRSSFQAFLE VVKSRSQPWE 600
601 TAEMDAIHSL QLILRDSFKE SEAAMNSKVV DGVVQPCRDM AGEQGIDELG AVAREMVRLI 660
661 ETATVPIFAV DAGGCINGWN AKIAELTGLS VEEAMGKSLV SDLIYKENEA TVNKLLSRAL 720
721 RGDEEKNVEV KLKTFSPELQ GKAVFVVVNA CSSKDYLNNI VGVCFVGQDV TSQKIVMDKF 780
781 INIQGDYKAI VHSPNPLIPP IFAADENTCC LEWNMAMEKL TGWSRSEVIG KMIVGEVFGS 840
841 CCMLKGPDAL TKFMIVLHNA IGGQDTDKFP FPFFDRNGKF VQALLTANKR VSLEGKVIGA 900
901 FCFLQIPSPE LQQALAVQRR QDTECFTKAK ELAYICQVIK NPLSGMRFAN SLLEATDLNE 960
961 DQKQLLETSV SCEKQISRIV GDMDLESIED GSFVLKREEF FLGSVINAIV SQAMFLLRDR1020
1021 GLQLIRDIPE EIKSIEVFGD QIRIQQLLAE FLLSIIRYAP SQEWVEIHLS QLSKQMADGF1080
1081 AAIRTEFRMA CPGEGLPPEL VRDMFHSSRW TSPEGLGLSV CRKILKLMNG EVQYIRESER1140
1141 SYFLIILELP VPRKRPLSTA SGSGDMMLMM PY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
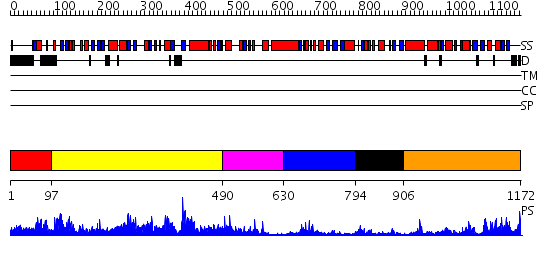
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..96] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [97..489] | 85.154902 | No description for 2oolA was found. | |
| 3 | View Details | [490..629] | 1.490979 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [630..793] | 6.69897 | Crystal Structure of the heme PAS sensor domain of Ec DOS (ferric form) | |
| 5 | View Details | [794..905] | 5.64 | No description for 2gj3A was found. | |
| 6 | View Details | [906..1172] | 36.69897 | Structure of the entire cytoplasmic portion of a sensor histidine kinase protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)