













| General Information: |
|
| Name(s) found: |
PSD3A_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 488 amino acids |
Gene Ontology: |
|
| Cellular Component: |
proteasome regulatory particle, lid subcomplex
[ISS]
plasma membrane [IDA] |
| Biological Process: |
embryonic development ending in seed dormancy
[NAS]
ubiquitin-dependent protein catabolic process [TAS] |
| Molecular Function: |
enzyme regulator activity
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTQDVEMKDN NTPSQSIISS STSTMQNLKE IAALIDTGSY TKEVRRIARA VRLTIGLRQK 60
61 LTGSVLSSFL DFALVPGSEA HSRLSSFVPK GDEHDMEVDT ASSATQAAPS KHLPAELEIY 120
121 CYFIVLLFLI DQKKYNEAKA CSSASIARLK NVNRRTIDVI ASRLYFYYSL SYEQTGDLAE 180
181 IRGTLLALHH SATLRHDELG QETLLNLLLR NYLHYNLYDQ AEKLRSKAPR FEAHSNQQFC 240
241 RYLFYLGKIR TIQLEYTDAK ESLLQAARKA PIAALGFRIQ CNKWAILVRL LLGEIPERSI 300
301 FTQKGMEKAL RPYFELTNAV RIGDLELFRT VQEKFLDTFA QDRTHNLIVR LRHNVIRTGL 360
361 RNISISYSRI SLPDVAKKLR LNSENPVADA ESIVAKAIRD GAIDATIDHK NGCMVSKETG 420
421 DIYSTNEPQT AFNSRIAFCL NMHNEAVRAL RFPPNTHKEK ESDEKRRERK QQEEELAKHM 480
481 AEEDDDDF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
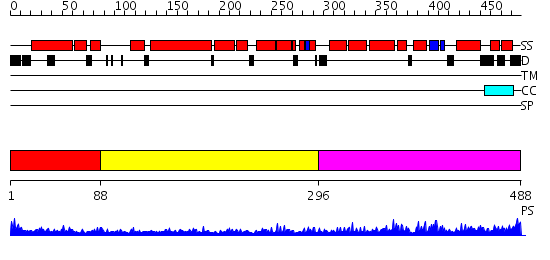
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | 1.228996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [88..295] | 4.69897 | Crystal structure of an 8 repeat consensus TPR superhelix (trigonal crystal form) | |
| 3 | View Details | [296..488] | 10.39794 | Solution structure of the PCI domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)