













| General Information: |
|
| Name(s) found: |
LOX2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 896 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA][ISS][TAS] chloroplast envelope [IDA] chloroplast thylakoid membrane [IDA] |
| Biological Process: |
jasmonic acid biosynthetic process
[IMP][TAS]
response to other organism [TAS] response to wounding [IEP] response to jasmonic acid stimulus [IEP] response to fungus [IEP] response to water deprivation [TAS] |
| Molecular Function: |
lipoxygenase activity
[IMP]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYCRESLSSL QTLNVAKSLS SLFPKQSALI NPISAGRRNN LPRPNLRRRC KVTASRANIE 60
61 QEGNTVKEPI QNIKVKGYIT AQEEFLEGIT WSRGLDDIAD IRGRSLLVEL ISAKTDQRIT 120
121 VEDYAQRVWA EAPDEKYECE FEMPEDFGPV GAIKIQNQYH RQLFLKGVEL KLPGGSITFT 180
181 CESWVAPKSV DPTKRIFFSD KSYLPSQTPE PLKKYRKEEL ETLQGKNREE VGEFTKFERI 240
241 YDYDVYNDVG DPDNDPELAR PVIGGLTHPY PRRCKTGRKP CETDPSSEQR YGGEFYVPRD 300
301 EEFSTAKGTS FTGKAVLAAL PSIFPQIESV LLSPQEPFPH FKAIQNLFEE GIQLPKDAGL 360
361 LPLLPRIIKA LGEAQDDILQ FDAPVLINRD RFSWLRDDEF ARQTLAGLNP YSIQLVEEWP 420
421 LISKLDPAVY GDPTSLITWE IVEREVKGNM TVDEALKNKR LFVLDYHDLL LPYVNKVREL 480
481 NNTTLYASRT LFFLSDDSTL RPVAIELTCP PNINKPQWKQ VFTPGYDATS CWLWNLAKTH 540
541 AISHDAGYHQ LISHWLRTHA CTEPYIIAAN RQLSAMHPIY RLLHPHFRYT MEINARARQS 600
601 LVNGGGIIET CFWPGKYALE LSSAVYGKLW RFDQEGLPAD LIKRGLAEED KTAEHGVRLT 660
661 IPDYPFANDG LILWDAIKEW VTDYVKHYYP DEELITSDEE LQGWWSEVRN IGHGDKKDEP 720
721 WWPVLKTQDD LIGVVTTIAW VTSGHHAAVN FGQYGYGGYF PNRPTTTRIR MPTEDPTDEA 780
781 LKEFYESPEK VLLKTYPSQK QATLVMVTLD LLSTHSPDEE YIGEQQEASW ANEPVINAAF 840
841 ERFKGKLQYL EGVIDERNVN ITLKNRAGAG VVKYELLKPT SEHGVTGMGV PYSISI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
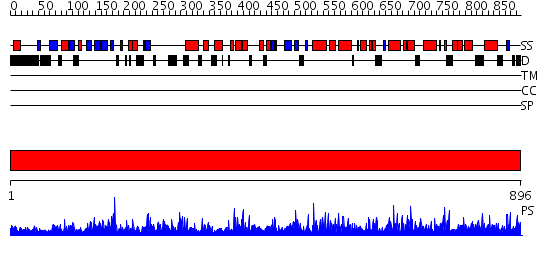
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..896] | 1000.0 | Lipoxigenase, C-terminal domain; Plant lipoxigenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)