













| General Information: |
|
| Name(s) found: |
GSA2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 472 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA][ISS] chloroplast envelope [IDA] |
| Biological Process: |
porphyrin biosynthetic process
[TAS]
|
| Molecular Function: |
pyridoxal phosphate binding
[IEA]
catalytic activity [IEA] transaminase activity [IEA] glutamate-1-semialdehyde 2,1-aminomutase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAATLTGSGI ALGFSCSAKF SKRASSSSNR RCIKMSVSVE EKTKKFTLQK SEEAFNAAKN 60
61 LMPGGVNSPV RAFKSVGGQP VVMDSAKGSR IRDIDGNEYI DYVGSWGPAI IGHADDEVLA 120
121 ALAETMKKGT SFGAPCLLEN VLAEMVISAV PSIEMVRFVN SGTEACMGVL RLARAFTGKQ 180
181 KFIKFEGCYH GHANSFLVKA GSGVATLGLP DSPGVPKAAT SDTLTAPYND IAAVEKLFEA 240
241 NKGEIAAIIL EPVVGNSGFI TPKPEFIEGI RRITKDNGAL LIFDEVMTGF RLAYGGAQEY 300
301 FGITPDLTTL GKIIGGGLPV GAYGGRRDIM EMVAPAGPMY QAGTLSGNPL AMTAGIHTLK 360
361 RLSQPGTYEY LDKITKELTN GILEAGKKTG HAMCGGYISG MFGFFFTEGP VYDFSDAKKS 420
421 DTEKFGKFFR GMLEEGVYLA PSQFEAGFTS LAHTSEDIQF TIAAAEKVLS RL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
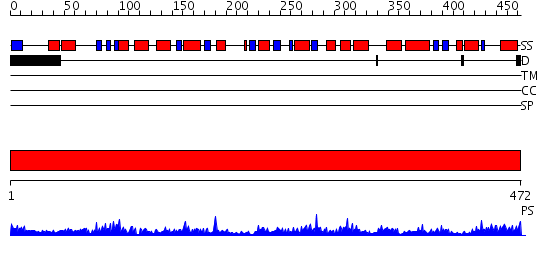
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..472] | 113.0 | Glutamate-1-semialdehyde aminomutase (aminotransferase) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|