













| General Information: |
|
| Name(s) found: |
G6PD2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 596 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[ISS]
|
| Biological Process: |
pentose-phosphate shunt, oxidative branch
[IDA]
glucose metabolic process [IDA][ISS] |
| Molecular Function: |
glucose-6-phosphate dehydrogenase activity
[IDA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAALSSSVTT RSYHSGYLAS FSPVNGDRHR SLSFLSASPQ GLNPLDLCVR FQRKSGRASV 60
61 FMQDGAIVTN SNSSESKTSL KGLKDEVLSA LSQEAAKVGV ESDGQSQSTV SITVVGASGD 120
121 LAKKKIFPAL FALYYEGCLP EHFTIFGYSR SKMTDVELRN MVSKTLTCRI DKRANCGEKM 180
181 EEFLKRCFYH SGQYDSQEHF TELDKKLKEH EAGRISNRLF YLSIPPNIFV DAVKCASTSA 240
241 SSVNGWTRVI VEKPFGRDSE TSAALTKSLK QYLEEDQIFR IDHYLGKELV ENLSVLRFSN 300
301 LIFEPLWSRQ YIRNVQFIFS EDFGTEGRGG YFDNYGIIRD IMQNHLLQIL ALFAMETPVS 360
361 LDAEDIRNEK VKVLRSMRPI RVEDVVIGQY KSHTKGGVTY PAYTDDKTVP KGSLTPTFAA 420
421 AALFIDNARW DGVPFLMKAG KALHTRSAEI RVQFRHVPGN LYNRNTGSDL DQATNELVIR 480
481 VQPDEAIYLK INNKVPGLGM RLDRSNLNLL YSARYSKEIP DAYERLLLDA IEGERRLFIR 540
541 SDELDAAWSL FTPLLKEIEE KKRIPEYYPY GSRGPVGAHY LAAKHKVQWG DVSIDQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
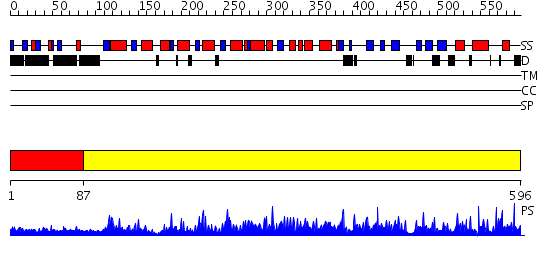
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..86] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [87..596] | 1000.0 | Glucose 6-phosphate dehydrogenase, N-terminal domain; Glucose 6-phosphate dehydrogenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)