













| General Information: |
|
| Name(s) found: |
DRL23_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 892 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
apoptosis
[IEA]
defense response [ISS] |
| Molecular Function: |
nucleotide binding
[IEA]
ATP binding [IEA] protein binding [IEA] nucleoside-triphosphatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSCISLQIS CDQVLTRAYS CFFSLGNYIH KLKDNIVALE KAIEDLTATR DDVLRRVQME 60
61 EGKGLERLQQ VQVWLKRVEI IRNQFYDLLS ARNIEIQRLC FYSNCSTNLS SSYTYGQRVF 120
121 LMIKEVENLN SNGFFEIVAA PAPKLEMRPI QPTIMGRETI FQRAWNRLMD DGVGTMGLYG 180
181 MGGVGKTTLL TQIHNTLHDT KNGVDIVIWV VVSSDLQIHK IQEDIGEKLG FIGKEWNKKQ 240
241 ESQKAVDILN CLSKKRFVLL LDDIWKKVDL TKIGIPSQTR ENKCKVVFTT RSLDVCARMG 300
301 VHDPMEVQCL STNDAWELFQ EKVGQISLGS HPDILELAKK VAGKCRGLPL ALNVIGETMA 360
361 GKRAVQEWHH AVDVLTSYAA EFSGMDDHIL LILKYSYDNL NDKHVRSCFQ YCALYPEDYS 420
421 IKKYRLIDYW ICEGFIDGNI GKERAVNQGY EILGTLVRAC LLSEEGKNKL EVKMHDVVRE 480
481 MALWTLSDLG KNKERCIVQA GSGLRKVPKV EDWGAVRRLS LMNNGIEEIS GSPECPELTT 540
541 LFLQENKSLV HISGEFFRHM RKLVVLDLSE NHQLDGLPEQ ISELVALRYL DLSHTNIEGL 600
601 PACLQDLKTL IHLNLECMRR LGSIAGISKL SSLRTLGLRN SNIMLDVMSV KELHLLEHLE 660
661 ILTIDIVSTM VLEQMIDAGT LMNCMQEVSI RCLIYDQEQD TKLRLPTMDS LRSLTMWNCE 720
721 ISEIEIERLT WNTNPTSPCF FNLSQVIIHV CSSLKDLTWL LFAPNITYLM IEQLEQLQEL 780
781 ISHAKATGVT EEEQQQLHKI IPFQKLQILH LSSLPELKSI YWISLSFPCL SGIYVERCPK 840
841 LRKLPLDSKT GTVGKKFVLQ YKETEWIESV EWKDEATKLH FLPSTKLVYI LS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
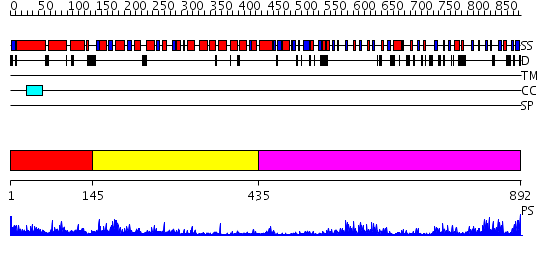
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..144] | 4.522879 | Crystal Structure Analysis of ClpB | |
| 2 | View Details | [145..434] | 5.221849 | Structure of the apoptotic protease-activating factor 1 bound to ADP | |
| 3 | View Details | [435..892] | 14.69897 | The molecular structure of toll-like receptor 3 ligand binding domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.77 |
Source: Reynolds et al. (2008)