













| General Information: |
|
| Name(s) found: |
DRL16_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 967 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
apoptosis
[IEA]
defense response [ISS] |
| Molecular Function: |
ATP binding
[IEA]
protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGNFVCIEIS GDQMLDRIIR CLCGKGYIRN LEKNLRALQR EMEDLRATQH EVQNKVAREE 60
61 SRHQQRLEAV QVWLDRVNSI DIECKDLLSV SPVELQKLCL CGLCSKYVCS SYKYGKRVFL 120
121 LLEEVTKLKS EGNFDEVSQP PPRSEVEERP TQPTIGQEEM LKKAWNRLME DGVGIMGLHG 180
181 MGGVGKTTLF KKIHNKFAET GGTFDIVIWI VVSQGAKLSK LQEDIAEKLH LCDDLWKNKN 240
241 ESDKATDIHR VLKGKRFVLM LDDIWEKVDL EAIGIPYPSE VNKCKVAFTT RDQKVCGQMG 300
301 DHKPMQVKCL EPEDAWELFK NKVGDNTLRS DPVIVGLARE VAQKCRGLPL ALSCIGETMA 360
361 SKTMVQEWEH AIDVLTRSAA EFSDMQNKIL PILKYSYDSL EDEHIKSCFL YCALFPEDDK 420
421 IDTKTLINKW ICEGFIGEDQ VIKRARNKGY EMLGTLIRAN LLTNDRGFVK WHVVMHDVVR 480
481 EMALWIASDF GKQKENYVVR ARVGLHEIPK VKDWGAVRRM SLMMNEIEEI TCESKCSELT 540
541 TLFLQSNQLK NLSGEFIRYM QKLVVLDLSH NPDFNELPEQ ISGLVSLQYL DLSWTRIEQL 600
601 PVGLKELKKL IFLNLCFTER LCSISGISRL LSLRWLSLRE SNVHGDASVL KELQQLENLQ 660
661 DLRITESAEL ISLDQRLAKL ISVLRIEGFL QKPFDLSFLA SMENLYGLLV ENSYFSEINI 720
721 KCRESETESS YLHINPKIPC FTNLTGLIIM KCHSMKDLTW ILFAPNLVNL DIRDSREVGE 780
781 IINKEKAINL TSIITPFQKL ERLFLYGLPK LESIYWSPLP FPLLSNIVVK YCPKLRKLPL 840
841 NATSVPLVEE FEIRMDPPEQ ENELEWEDED TKNRFLPSIK PLVRRLKIHY SGMGFLNVKN 900
901 QNPRFFFYCF IYLLVVHLDC IIDLHSDTSG MCCVVHLDYV FHFPFVFPKT FCILFRLHFY 960
961 TIKSLCV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
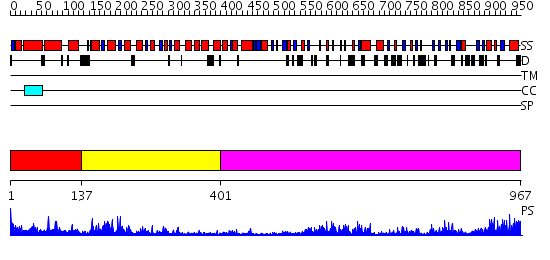
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..136] | 3.715982 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [137..400] | 8.30103 | Structure of the apoptotic protease-activating factor 1 bound to ADP | |
| 3 | View Details | [401..967] | 20.0 | The molecular structure of toll-like receptor 3 ligand binding domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)