













| General Information: |
|
| Name(s) found: |
DPNP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 353 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
nucleus [IDA] chloroplast [IEP] cytoplasm [IDA] |
| Biological Process: |
sulfur metabolic process
[IGI][ISS]
regulation of jasmonic acid biosynthetic process [IMP] response to cation stress [IGI] response to light stimulus [IEP][IMP] RNA catabolic process [IMP] response to water deprivation [IMP] photoperiodism, flowering [IMP] response to cold [IMP] response to abiotic stimulus [IMP] abscisic acid mediated signaling pathway [IMP] negative regulation of signal transduction [IMP] phosphoinositide-mediated signaling [TAS] negative regulation of transcription [IEP] miRNA catabolic process [IMP] positive regulation of unidimensional cell growth [IMP] |
| Molecular Function: |
inositol or phosphatidylinositol phosphatase activity
[IDA][ISS]
3'(2'),5'-bisphosphate nucleotidase activity [IDA][ISS] nucleotide phosphatase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAYEKELDAA KKAASLAARL CQKVQKALLQ SDVQSKSDKS PVTVADYGSQ AVVSLVLEKE 60
61 LSSEPFSLVA EEDSGDLRKD GSQDTLERIT KLVNDTLATE ESFNGSTLST DDLLRAIDCG 120
121 TSEGGPNGRH WVLDPIDGTK GFLRGDQYAV ALGLLEEGKV VLGVLACPNL PLASIAGNNK 180
181 NKSSSDEIGC LFFATIGSGT YMQLLDSKSS PVKVQVSSVE NPEEASFFES FEGAHSLHDL 240
241 SSSIANKLGV KAPPVRIDSQ AKYGALSRGD GAIYLRFPHK GYREKIWDHV AGAIVVTEAG 300
301 GIVTDAAGKP LDFSKGKYLD LDTGIIVANE KLMPLLLKAV RDSIAEQEKA SAL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
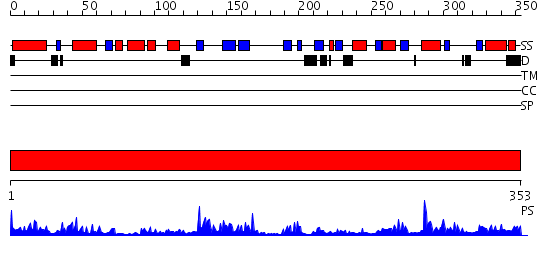
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..353] | 61.39794 | 3';5'-adenosine bisphosphatase, PAP phosphatase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)