













| General Information: |
|
| Name(s) found: |
DNLI1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 790 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
mitochondrion [IDA] |
| Biological Process: |
DNA repair
[IEA]
DNA replication [IEA] DNA recombination [IEA] |
| Molecular Function: |
ATP binding
[IEA][ISS]
DNA binding [IEA] DNA ligase (ATP) activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLAIRSSNYL RCIPSLCTKT QISQFSSVLI SFSRQISHLR LSSCHRAMSS SRPSAFDALM 60
61 SNARAAAKKK TPQTTNLSRS PNKRKIGETQ DANLGKTIVS EGTLPKTEDL LEPVSDSANP 120
121 RSDTSSIAED SKTGAKKAKT LSKTDEMKSK IGLLKKKPND FDPEKMSCWE KGERVPFLFV 180
181 ALAFDLISNE SGRIVITDIL CNMLRTVIAT TPEDLVATVY LSANEIAPAH EGVELGIGES 240
241 TIIKAISEAF GRTEDHVKKQ NTELGDLGLV AKGSRSTQTM MFKPEPLTVV KVFDTFRQIA 300
301 KESGKDSNEK KKNRMKALLV ATTDCEPLYL TRLLQAKLRL GFSGQTVLAA LGQAAVYNEE 360
361 HSKPPPNTKS PLEEAAKIVK QVFTVLPVYD IIVPALLSGG VWNLPKTCNF TLGVPIGPML 420
421 AKPTKGVAEI LNKFQDIVFT CEYKYDGERA QIHFMEDGTF EIYSRNAERN TGKYPDVALA 480
481 LSRLKKPSVK SFILDCEVVA FDREKKKILP FQILSTRARK NVNVNDIKVG VCIFAFDMLY 540
541 LNGQQLIQEN LKIRREKLYE SFEEDPGYFQ FATAVTSNDI DEIQKFLDAS VDVGCEGLII 600
601 KTLDSDATYE PAKRSNNWLK LKKDYMDSIG DSVDLVPIAA FHGRGKRTGV YGAFLLACYD 660
661 VDKEEFQSIC KIGTGFSDAM LDERSSSLRS QVIATPKQYY RVGDSLNPDV WFEPTEVWEV 720
721 KAADLTISPV HRAATGIVDP DKGISLRFPR LLRVREDKKP EEATSSEQIA DLYQAQKHNH 780
781 PSNEVKGDDD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
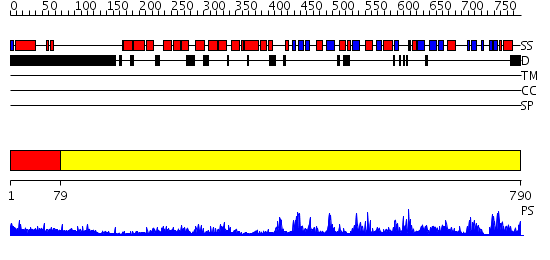
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..78] | 3.198997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [79..790] | 177.0 | Crystal Structure of Human DNA Ligase I bound to 5'-adenylated, nicked DNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)