













| General Information: |
|
| Name(s) found: |
DHE1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 411 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA]
|
| Biological Process: |
response to salt stress
[IEP]
nitrogen compound metabolic process [IMP] response to cadmium ion [IEP] response to absence of light [IEP] |
| Molecular Function: |
ATP binding
[IDA]
zinc ion binding [IDA] copper ion binding [IDA] oxidoreductase activity [ISS] glutamate dehydrogenase [NAD(P)+] activity [IDA][IMP] cobalt ion binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNALAATNRN FKLAARLLGL DSKLEKSLLI PFREIKVECT IPKDDGTLAS FVGFRVQHDN 60
61 ARGPMKGGIR YHPEVDPDEV NALAQLMTWK TAVAKIPYGG AKGGIGCDPS KLSISELERL 120
121 TRVFTQKIHD LIGIHTDVPA PDMGTGPQTM AWILDEYSKF HGYSPAVVTG KPIDLGGSLG 180
181 RDAATGRGVM FGTEALLNEH GKTISGQRFV IQGFGNVGSW AAKLISEKGG KIVAVSDITG 240
241 AIKNKDGIDI PALLKHTKEH RGVKGFDGAD PIDPNSILVE DCDILVPAAL GGVINRENAN 300
301 EIKAKFIIEA ANHPTDPDAD EILSKKGVVI LPDIYANSGG VTVSYFEWVQ NIQGFMWEEE 360
361 KVNDELKTYM TRSFKDLKEM CKTHSCDLRM GAFTLGVNRV AQATILRGWG A |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
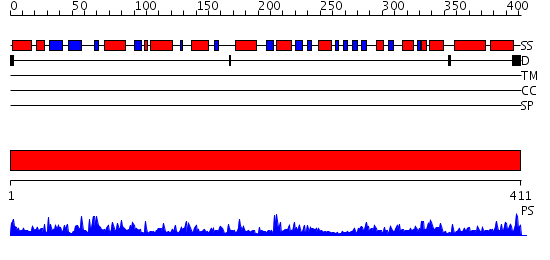
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..411] | 128.0 | Glutamate dehydrogenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)