













| General Information: |
|
| Name(s) found: |
CATA3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 492 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
peroxisome [IDA][TAS] chloroplast [IDA] cytosolic ribosome [IDA] cell wall [IDA] membrane [IDA] apoplast [IDA] mitochondrion [IDA] chloroplast envelope [IDA] vacuole [IDA] plasma membrane [IDA] |
| Biological Process: |
response to cold
[IEP]
cellular response to sulfate starvation [IEP] response to cadmium ion [IEP] cellular response to phosphate starvation [IEP] response to light stimulus [IEP] cellular response to nitrogen starvation [IEP] hydrogen peroxide catabolic process [TAS] |
| Molecular Function: |
catalase activity
[ISS]
cobalt ion binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDPYKYRPSS AYNAPFYTTN GGAPVSNNIS SLTIGERGPV LLEDYHLIEK VANFTRERIP 60
61 ERVVHARGIS AKGFFEVTHD ISNLTCADFL RAPGVQTPVI VRFSTVVHER ASPETMRDIR 120
121 GFAVKFYTRE GNFDLVGNNT PVFFIRDGIQ FPDVVHALKP NPKTNIQEYW RILDYMSHLP 180
181 ESLLTWCWMF DDVGIPQDYR HMEGFGVHTY TLIAKSGKVL FVKFHWKPTC GIKNLTDEEA 240
241 KVVGGANHSH ATKDLHDAIA SGNYPEWKLF IQTMDPADED KFDFDPLDVT KIWPEDILPL 300
301 QPVGRLVLNR TIDNFFNETE QLAFNPGLVV PGIYYSDDKL LQCRIFAYGD TQRHRLGPNY 360
361 LQLPVNAPKC AHHNNHHEGF MNFMHRDEEI NYYPSKFDPV RCAEKVPTPT NSYTGIRTKC 420
421 VIKKENNFKQ AGDRYRSWAP DRQDRFVKRW VEILSEPRLT HEIRGIWISY WSQADRSLGQ 480
481 KLASRLNVRP SI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
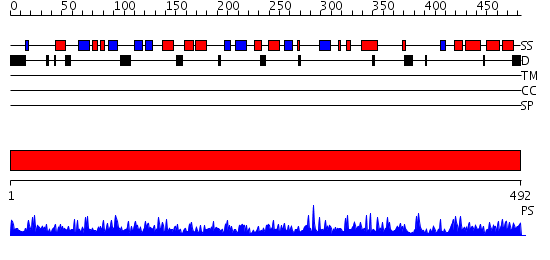
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..492] | 1000.0 | The crystal structure of bovine liver catalase without NADPH |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)