













| General Information: |
|
| Name(s) found: |
AS1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 367 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
response to salt stress
[IEP]
asymmetric cell division [IMP] response to jasmonic acid stimulus [IEP] response to cadmium ion [IEP] response to salicylic acid stimulus [IEP] response to virus [IMP] polarity specification of adaxial/abaxial axis [IMP] defense response to bacterium [IMP] leaf morphogenesis [IMP] response to auxin stimulus [IEP] response to gibberellin stimulus [IEP] negative regulation of transcription [IDA] regulation of innate immune response [IMP] leaf formation [IGI] defense response to fungus [IMP] |
| Molecular Function: |
DNA binding
[IPI][ISS]
transcription factor activity [ISS] protein homodimerization activity [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKERQRWSGE EDALLRAYVR QFGPREWHLV SERMNKPLNR DAKSCLERWK NYLKPGIKKG 60
61 SLTEEEQRLV IRLQEKHGNK WKKIAAEVPG RTAKRLGKWW EVFKEKQQRE EKESNKRVEP 120
121 IDESKYDRIL ESFAEKLVKE RSNVVPAAAA AATVVMANSN GGFLHSEQQV QPPNPVIPPW 180
181 LATSNNGNNV VARPPSVTLT LSPSTVAAAA PQPPIPWLQQ QQPERAENGP GGLVLGSMMP 240
241 SCSGSSESVF LSELVECCRE LEEGHRAWAD HKKEAAWRLR RLELQLESEK TCRQREKMEE 300
301 IEAKMKALRE EQKNAMEKIE GEYREQLVGL RRDAEAKDQK LADQWTSRHI RLTKFLEQQM 360
361 GCRLDRP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
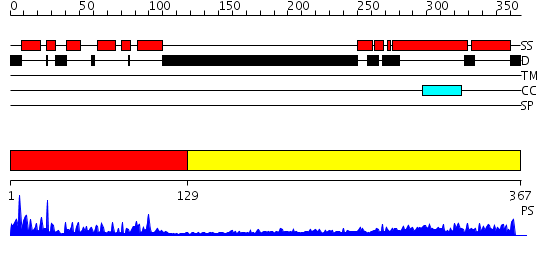
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..128] | 33.69897 | CHICKEN B-MYB DNA BINDING DOMAIN, REPEAT 2 AND REPEAT3, NMR, 32 STRUCTURES | |
| 2 | View Details | [129..367] | 1.16 | crystal structure of the cytotoxic bacterial protein colicin B at 2.5 A resolution |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)