













| General Information: |
|
| Name(s) found: |
zag-1 /
CE34546
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 596 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC][IEA intracellular [IEA |
| Biological Process: |
locomotion
[IMP]
negative regulation of transcription [TAS axon guidance [IMP regulation of transcription, DNA-dependent [IEA positive regulation of axon extension [IMP |
| Molecular Function: |
nucleic acid binding
[IEA transcription factor activity [IEA zinc ion binding [IEA sequence-specific DNA binding [IEA transcription repressor activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVDIAEAMPT TASSLPSDEA LRKFKCPECT KAFKFKHHLK EHIRIHSGEK PFECQQCHKR 60
61 FSHSGSYSSH MSSKKCVQQA SPSMVTPFNP YQLMMYRNIM LQLQTPQVSF LPSTAANNMD 120
121 YMSLLQANLF QSLENGTSPT PTQEPSAPAS PEPKIEVVDE PEVSSEVKTE VKTEVKTEDS 180
181 VPEESITPAV SMSLSPAPEQ NGNESMNNGG SGSDGKSSPD WRPLRSRSFL NDSQVAVLQN 240
241 HFKRNPFPSK YELSAVAEQI GVNKRVVQVW FQNTRAKERR SNRLPSMPRG SVASAAAAAA 300
301 TSPTVWQTPV QLMAAWASQF SNGNNSLTAS QDERNNENTD EVMDHDGLKD GKETPLDLTL 360
361 STDDTEPEWS PEKLIGFLDQ TGGVIQELLR QAGNGFVTNQ EDEEEKPIKA EESPVSSGSS 420
421 SIWPSFIGQY PSILDSASLS VLEKALDQQK SSEDDASSLC SNESKLLKFP TTPLKEEEGL 480
481 FSCDQCDKVF GKQSSLARHK YEHSGQRPYK CDICEKAFKH KHHLTEHKRL HSGEKPFQCD 540
541 KCLKRFSHSG SYSQHMNHRY SYCKPYREQP ASPSDVLNGG SVTVSPSSSN TPPPST |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
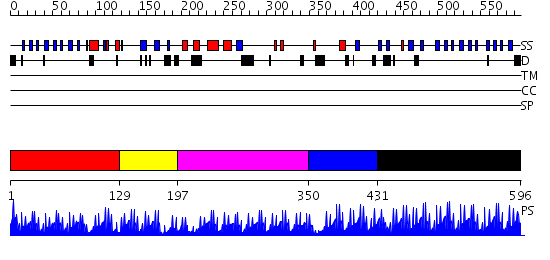
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..128] | 4.045757 | No description for 2dlkA was found. | |
| 2 | View Details | [129..196] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [197..349] | 17.69897 | Transcription factor IIIA, TFIIIA | |
| 4 | View Details | [350..430] | 0.983 | SOLUTION STRUCTURE OF THE MU NER PROTEIN BY MULTIDIMENSIONAL NMR | |
| 5 | View Details | [431..596] | 57.154902 | No description for 2i13A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)