













| General Information: |
|
| Name(s) found: |
unc-62 /
CE33584
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 523 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
oocyte construction
[IMP locomotory behavior [IMP hermaphrodite genitalia development [IMP embryonic development ending in birth or egg hatching [IMP regulation of transcription, DNA-dependent [IEA oviposition [IMP cell migration [IMP positive regulation of multicellular organism growth [IMP |
| Molecular Function: |
transcription factor activity
[IEA sequence-specific DNA binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAQRQFNDGY GPPPGSASAD PASYIADPAA FYNLYTNMGG APTSTPMMHH EMGEAMKRDK 60
61 ESIYAHPLYP LLVLLFEKCE LATSTPRDTS RDGSTSSDVC SSASFKDDLN EFVRHTQENA 120
121 DKQYYVPNPQ LDQIMLQSIQ MLRFHLLELE KVHELCDNFC NRYVVCLKGK MPLDIVGDER 180
181 ASSSQPPMSP GSMGHLGHSS SPSMAGGATP MHYPPPYEPQ SVPLPENAGV MGGHPMEGSS 240
241 MAYSMAGMAA AAASSSSSSN QAGDHPLANG GTLHSTAGAS QTLLPIAVSS PSTCSSGGLR 300
301 QDSTPLSGET PMGHANGNSM DSISEAAPSS ASSSSSSKNK PSTPNGGRIG KSRGRGIFPK 360
361 QATNRLRQWL FQNLTHPYPS EEQKKQLAKE TGLTILQVNN WFINARRRIV QPMIDQNNRA 420
421 GRSGQMNVCK NRRRNRSEQS PGPSPDSGSD SGANYSPDPS SLAASTAMPY PAEFYMQRTM 480
481 PYGGFPSFTN PAMPFMNPMM GFQVAPTVDA LSQQWVDLSA PHE |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
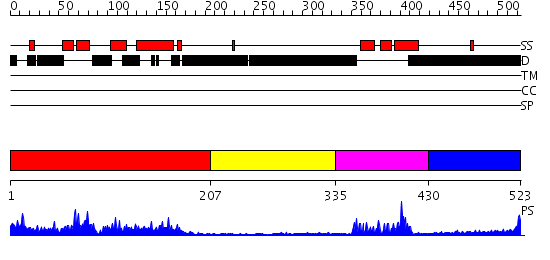
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..206] | 5.205976 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [207..334] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [335..429] | 11.69897 | Solution structure of the homeobox domain of human homeobox protein PKNOX1 | |
| 4 | View Details | [430..523] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)