













| General Information: |
|
| Name(s) found: |
lin-40 /
CE33330
[WormBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 756 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
negative regulation of vulval development
[IMP locomotory behavior [IMP hermaphrodite genitalia development [IMP nematode larval development [IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP post-embryonic body morphogenesis [IMP regulation of transcription, DNA-dependent [IEA oviposition [IMP gamete generation [IMP |
| Molecular Function: |
DNA binding
[IEA transcription factor activity [IEA zinc ion binding [IEA sequence-specific DNA binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSSSLSSTS TRVTYAVGDF VYFDDTSATD APYQIRKIEE LVKTEKGGVD ARCVVYLRRR 60
61 DIPQHLLKIA DQAQRRFDNY YEVDKKKPEN FTTKGFIVVN GASEAAPEAA PEATEASAET 120
121 SETKVKEDTD DVAEKMEEDE ECPAPVGQGR TSSTEEPSAS DAPAATAKDK KEEKEAKENA 180
181 KNLKETAEAE ATKLIDWGDG GLPLGVDKLT PDQRLKLRQH EIFMTRQSEI LPAAAIRGKC 240
241 RVVLLGDGEE AQNYLPLDDT FYHSLVYDPN AQTLLADKGA IRVGEKYQAV VDEWMEPADR 300
301 EAKEAKVLAE KVKKEEEEAK LKAEAEEDTE NDGLTIAEDE EMPELKNEEK EETKVEEEAT 360
361 TTDSEGEVGE VLVWHPHHAL TDRDIDQYMI VARSVGLFAR AIDGASAPKL PTLQLAAAFA 420
421 SRDVTIFHAH AILHQANYDV GQAVKYLVPV ASRDTYPCQV DDSSGLQTKT LGGPILCRDQ 480
481 LEEWSTPEMN LFEDALDKVG KDFNEIRAEY LPWKSIRDIV EYYYLMKASN RYTDRKKNKP 540
541 SAGGGSSTTT DEKFTNIYIP PFNKPIAASI LPYNTSQTTL MNNDNPCENC GTLDALNWYQ 600
601 WGGVGDKKVL CSTCWIQWKK FAGLNQKHEL ERFDKTRPPP VEPVAPTPAA SNGTPASQKA 660
661 AATAAVNTLT PQQQQQQRAA AANMLMTLNS GDKQNLLNVA KEALARGQIT QEQFLQLQKN 720
721 FAQKIQAQIQ QQIQQQQQRQ AQTTPQVVAT PRVRIG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
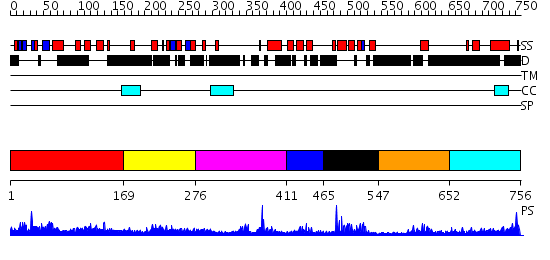
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..168] | 2.03095 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [169..275] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [276..410] | 11.49485 | No description for PF01448.15 was found. No confident structure predictions are available. | |
| 4 | View Details | [411..464] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [465..546] | 15.0 | Solution structure of the myb-like DNA-binding domain of mouse MTA3 protein | |
| 6 | View Details | [547..651] | 6.522879 | Erythroid transcription factor GATA-1 | |
| 7 | View Details | [652..756] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 |
|
|||||||||
| 6 |
|
|||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.74 |
Source: Reynolds et al. (2008)