













| General Information: |
|
| Name(s) found: |
CE33172
[WormBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 695 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA intracellular [IEA |
| Biological Process: |
mRNA processing
[IEA reproduction [IMP |
| Molecular Function: |
nucleic acid binding
[IEA zinc ion binding [IEA RNA binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPNDVDNTKI TLSLGKCGYL PQDVRTMTKF DKNTKQIRTF AFVEFSNRET AEQWMTDYEG 60
61 WLTLDDGRTL AVEYAKGDPA AGSGNKGDDW ICAHCSMNNF VKRQTCFKCE ISKDQSMELE 120
121 KLGAHMVGMT PCDTLLIRGL PDGINNSLIF DSLGALVCLS SISMIKLSES KRFAYLQMKS 180
181 ADEAKMLLNL TYKSPIRIKD KDVQVSWCRD SMSKLIQQQM TMLTAGPGKN SVATGSLAQG 240
241 NMTGAEIAAA ALSKANAVRQ ASQQVGQIMT GLPAPAANMI PPNFSVPPPN LSVPPPMQAT 300
301 QPEHQNGVIG MTQTPKGLLP RYLPPNPLTF AHDPNLGYYV DPITKFCYDS ATGYYFNNAT 360
361 SQWCNWDLTH HTYFPVETPA VINSADPEEE KMKKNEEGPK TAQDLVKDMA KWAKRQEKDK 420
421 KKVQISIKGK ETKGIELKNV FSNEKQKRIA QTAALFDDDE EEDQEEEVRG RRSEEPSTSS 480
481 SFSSVPLAPR KSTLQDVRDA MERALYDETK KTCMLCKRAF SDVEVLRKHV EKSDLHRNNL 540
541 EAKRAEWGRE TAAKLQEEEE DASAPDLPKI VYRDRAKERR RQFGIDSTGY AFDVMGGPPG 600
601 PTSVRHEETI RKVSEEASKR PLDESNIGNR LLKSMGWKEG QGVGKHAQGI VNPIEAERFV 660
661 QGAGLGAAGS KMRHGAEASH KEKTRQALYS RYHDA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
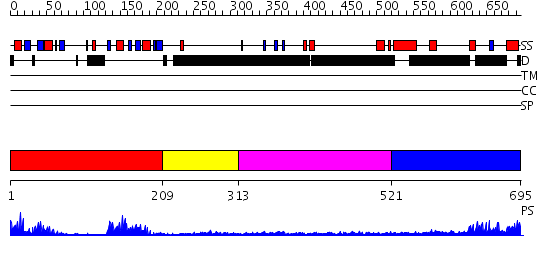
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..208] | 27.69897 | Crystal Structure of UP1 Complexed With d(TAGG(6MI)TTAGGG): A Human Telomeric Repeat Containing 6-methyl-8-(2-deoxy-beta-ribofuranosyl)isoxanthopteridine (6MI) | |
| 2 | View Details | [209..312] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [313..520] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [521..695] | 16.147993 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)