













| General Information: |
|
| Name(s) found: |
hst-1 /
CE33047
[WormBase]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 715 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
embryonic development ending in birth or egg hatching
[IMP reproduction [IMP |
| Molecular Function: |
sulfotransferase activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAQWNRQLLD KYCKEYRVPM FSFMSSKPND QLKRIKIKGS SLWMWQNQRI QRLAVTPSII 60
61 HRISKIGNYR QFSSSDPADW ILFETSEKFE SVLSGTVKSG YERAVVVRDK GLEDGVERII 120
121 FGRNLTDFQV KITFLDALWW AMGNQKSFTL DRFVQVDIDD VFVGAQGTRI VEEDVRKLIS 180
181 TQKEFRNYVQ NFTFMLGFSG SYFRNGDDLE DRGDEFLVEN AEKFVWFPHM WRHNHAHEHN 240
241 FTYLEAIMAQ NKLFAQNMHL PVDYPYAIAP QHDGVFPVHE QLFRAWRKVW NVSVTATEEY 300
301 PHFKPATARK GFIHAGIHVL PRQTCGLYTH TQLFDEYPEG FDKVQKSIEG GDLFFTILLN 360
361 PISIFMTHQQ NYAYDRLALY TFENLFRFIK CWTNIKLKWQ DPLTSSQLYF QKFPDERTPL 420
421 WTNPCTDPRH HAILPPSINC TKKSLPDLLI IGPQKTGSTA LASFLSLHPN TSQNTPVPGS 480
481 FEEVQFFGGQ NYLKGVEWYM SNFPSSSTVT FEKSATYFDN PSAPKQAASL VPHAKIVIIL 540
541 QNPAQRAYSW FQHILAHEDP VAITAGSLEV ILDSNSTSSK KVRQRCISGG RYVHHLTKWL 600
601 EHFSLQQMIF VDSDELKMKP PTVLNSLSKW LDLPEFPFET YIRYSPSKGF HCRLLDGKTK 660
661 CLGESKGRKY PEMPENLRRK LDKIFSLDNS ALYKFLRKNR LKIPTWLEES VRIRA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
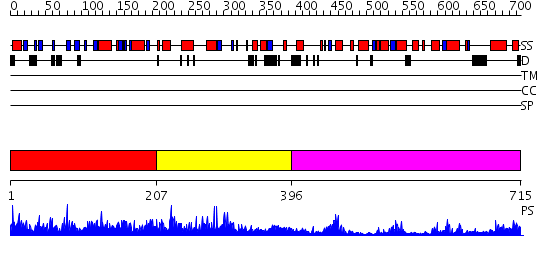
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..206] | 1.020915 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [207..395] | 1.056921 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [396..715] | 60.69897 | Heparan sulfate N-deacetylase/N-sulfotransferase domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)