













| General Information: |
|
| Name(s) found: |
CE32815
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 504 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
aromatic amino acid family metabolic process
[IEA cellular amino acid and derivative metabolic process [IEA biosynthetic process [IEA histidine biosynthetic process [IEA |
| Molecular Function: |
pyridoxal phosphate binding
[IEA transferase activity, transferring nitrogenous groups [IEA L-tyrosine:2-oxoglutarate aminotransferase activity [IEA transaminase activity [IEA histidinol-phosphate transaminase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRTVQAISGL VTSRFFGTST RIMASGKTLN TSNINPNVIK MEYAVRGPIV IRAVELEKEL 60
61 ATGAQKPFPN VIKANIGDAH AMGQKPITFI RQLLACIVNP EIMKTDKSIP SDVIEHANAF 120
121 LGSCGGKSAG AYSQSTGVEI VRKHVAEYIK RRDGGIPCNS EDVCLSGGAS ESIRNVLKLF 180
181 INHNNAKKVG VMIPIPQYPL YSATIEEFGL GQVGYYLSES SNWSMDEAEL ERSFNDHCKE 240
241 YDIRVLCIIN PGNPTGQALS RENIETIIKF AQKKNLFLMA DEVYQDNVYA QGSQFHSFKK 300
301 VLVEMGEPYN KMELASFHSV SKGYMGECGM RGGYVEFLNL DPEVYVLFKK MISAKLCSTV 360
361 LGQAVIDAVV NPPKEGDASY ALWKQEKDAV LASLKERATL VEKAYSSIDG ISCNPVQGAM 420
421 YAFPQITIPQ RAVEKAQSLN QQPDFFYAMQ LLETTGICIV PGSGFGQKDG TYHFRTTILP 480
481 QPELFKDMLS RFTDFHQKFL AEYK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
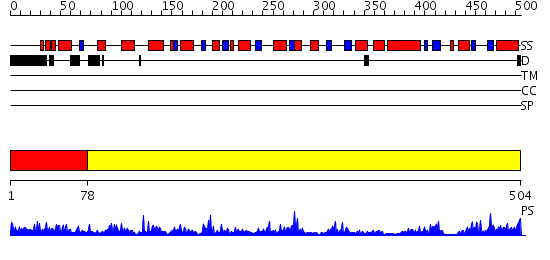
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..77] | 3.154902 | No description for 2ra5A was found. | |
| 2 | View Details | [78..504] | 78.221849 | Alanine aminotransferase from Pyrococcus furiosus Pfu-1397077-001 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.63 |
Source: Reynolds et al. (2008)