













| General Information: |
|
| Name(s) found: |
aco-2 /
CE32436
[WormBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 683 amino acids |
Gene Ontology: |
|
| Cellular Component: |
3-isopropylmalate dehydratase complex
[IEA |
| Biological Process: |
lysine biosynthetic process
[IEA locomotory behavior [IMP hermaphrodite genitalia development [IMP nematode larval development [IMP embryonic development ending in birth or egg hatching [IMP metabolic process [IEA leucine biosynthetic process [IEA tricarboxylic acid cycle [IEA cellular amino acid biosynthetic process [IEA reproduction [IMP growth [IMP |
| Molecular Function: |
hydro-lyase activity
[IEA homoaconitate hydratase activity [IEA 4 iron, 4 sulfur cluster binding [IEA 3-isopropylmalate dehydratase activity [IEA aconitate hydratase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQDATAQMAM LQFISSGLPK TAVPSTIHCD HLIEAQKGGA QDLARAKDLN KEVFNFLATA 60
61 GSKYGVGFWK PGSGIIHQII LENYAFPGLL LIGTDSHTPN GGGLGGLCIG VGGADAVDVM 120
121 ADIPWELKCP KVIGIKLTGK LNGWTSAKDV ILKVADILTV KGGTGAIVEY FGPGVDSISA 180
181 TGMGTICNMG AEIGATTSVF PYNESMYKYL EATGRKEIAE EARKYKDLLT ADDGANYDQI 240
241 IEINLDTLTP HVNGPFTPDL ASSIDKLGEN AKKNGWPLDV KVSLIGSCTN SSYEDMTRAA 300
301 SIAKQALDKG LKAKTIFTIT PGSEQVRATI ERDGLSKIFA DFGGMVLANA CGPCIGQWDR 360
361 QDVKKGEKNT IVTSYNRNFT GRNDANPATH GFVTSPDITT AMAISGRLDF NPLTDELTAA 420
421 DGSKFKLQAP TGLDLPPKGY DPGEDTFQAP SGSGQVDVSP SSDRLQLLSP FDKWDGKDLE 480
481 DMKILIKVTG KCTTDHISAA GPWLKYRGHL DNISNNLFLT AINADNGEMN KVKNQVTGEY 540
541 GAVPATARKY KADGVRWVAI GDENYGEGSS REHAALEPRH LGGRAIIVKS FARIHETNLK 600
601 KQGMLPLTFA NPADYDKIDP SDNVSIVGLS SFAPGKPLTA IFKKTNGSKV EVTLNHTFNE 660
661 QQIEWFKAGS ALNRMKEVFA KSK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
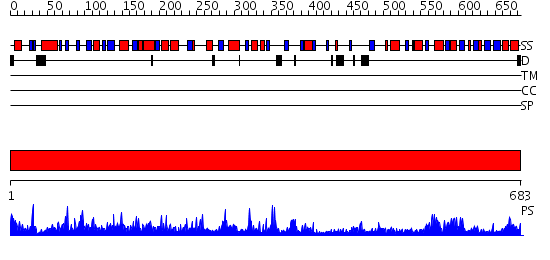
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..683] | 1000.0 | STRUCTURE OF ACTIVATED ACONITASE. FORMATION OF THE (4FE-4S) CLUSTER IN THE CRYSTAL |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)