













| General Information: |
|
| Name(s) found: |
wah-1 /
CE31841
[WormBase]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 700 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mitochondrion [IDA cytosol [IDA |
| Biological Process: |
electron transport
[IEA cell death [IMP nematode larval development [IMP DNA fragmentation involved in apoptosis [IDA glutathione metabolic process [IEA embryonic development ending in birth or egg hatching [IMP positive regulation of growth rate [IMP growth [IMP reproduction [IMP detoxification of mercury ion [IEA |
| Molecular Function: |
NADP or NADPH binding
[IEA oxidoreductase activity [IEA FAD binding [IEA mercury ion binding [IEA mercury (II) reductase activity [IEA glutathione-disulfide reductase activity [IEA dihydrolipoyl dehydrogenase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLLRAVGRQM TSVIFRQQTA VRSIAMSRVA LGGGGHHHEP TPVYIPKPGS LDWTFFSRSH 60
61 TKSAHEFEPY KPEIGAFIGA VAFIGLTLIA VVIKTDVFKK EDSHGHGHGH AKHSKKHEEK 120
121 HEQKHEEKEH AEPEKKEEAK PEKPAEPKEP EPAQKQAEQP EQAEEKQETK DAEPKEQVDD 180
181 RQTEEAVHAR RAPAAAEEPA PSTSKADAVE EKRSEQQSMK PSESTEENTT TTSADGLLHC 240
241 EYVIIGSGTA AYYASLSIRA KQAEAKVLMI GEEPELPYNR PPLSKELWWY GDETSATKLA 300
301 YTPLSGKKRD IFYEVDGFFV SPEDLPKAVH GGVALLRGRK AVKICEEDKK VILEDGTTIG 360
361 YDKLLIATGV RPKKEQVFEE ASEEAKQKIT YFHYPADFKR VERGLADKSV QKVTIIGNGL 420
421 LASELSYSIK RKYGENVEVH QVFEEKYPAE DILPEHIAQK SIEAIRKGGV DVRAEQKVEG 480
481 VRKCCKNVVL KLSDGSELRT DLVVVATGEE PNSEIIEASG LKIDEKLGGV RADKCLKVGE 540
541 NVWAAGAIAT FEDGVLGARR VSSWENAQIS GRLAGENMAT AAADGKSEGK AFWYQPSFFT 600
601 KFAPHLHINA IGKCDSSLET VSVHAEPDKD TPLEKAVVFY KSKEDGSIVG VLLLNVFGPS 660
661 LDVARRIIDD RKKVDEYKEI AKLFPLYDPV KSDEDDAKSA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
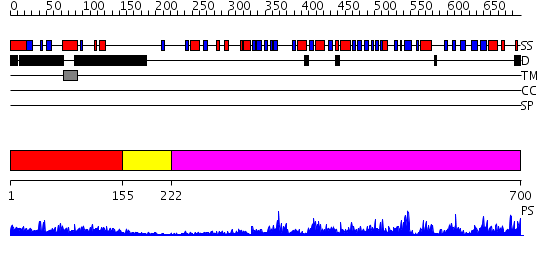
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..154] | 1.133996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [155..221] | 9.30103 | Trimethylamine dehydrogenase, N-terminal domain; Trimethylamine dehydrogenase, C-terminal domain; Trimethylamine dehydrogenase, middle domain | |
| 3 | View Details | [222..700] | 55.045757 | Apoptosis-inducing factor (AIF) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|