













| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYTDSNNRNF DEVNHQHQQE QDFNGQSKYD YPQFNRPMGL RWRDDQRMME YFMSNGPVET 60
61 VPVMPILTEH PPASPFGRGP STERPTTSSR YEYSSPSLED IDLIDVLWRS DIAGEKGTRQ 120
121 VAPADQYECD LQTLTEKSTV APLTAEENAR YEDLSKGFYN GFFESFNNNQ YQQKHQQQQR 180
181 EQIKTPTLEH PTQKAELEDD LFDEDLAQLF EDVSREEGQL NQLFDNKQQH PVINNVSLSE 240
241 GIVYNQANLT EMQEMRDSCN QVSISTIPTT STAQPETLFN VTDSQTVEQW LPTEVVPNDV 300
301 FPTSNYAYIG MQNDSLQAVV SNGQIDYDHS YQSTGQTPLS PLIIGSSGRQ QQTQTSPGSV 360
361 TVTATATQSL FDPYHSQRHS FSDCTTDSSS TCSRLSSESP RYTSESSTGT HESRFYGKLA 420
421 PSSGSRYQRS SSPRSSQSSI KIARVVPLAS GQRKRGRQSK DEQLASDNEL PVSAFQISEM 480
481 SLSELQQVLK NESLSEYQRQ LIRKIRRRGK NKVAARTCRQ RRTDRHDKMS HYI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
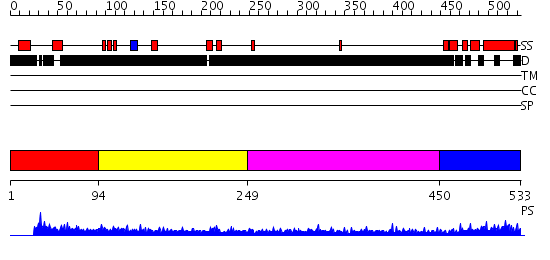
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..93] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [94..248] | 2.159996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [249..449] | 4.165982 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [450..533] | 19.221849 | Skn-1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)