













| General Information: |
|
| Name(s) found: |
CE31475
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 729 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
positive regulation of growth rate
[IMP ATP metabolic process [IEA nucleobase, nucleoside, nucleotide and nucleic acid metabolic process [IEA |
| Molecular Function: |
ATP binding
[IEA adenylate kinase activity [IEA nucleotide kinase activity [IEA phosphotransferase activity, phosphate group as acceptor [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGPSEARIY LQDHRIPQLF EGLMTGLIYS RPADPLKFLE ESIVKIRSTP GITLSWDMFI 60
61 GPEAGTQGRQ YPPGAPHLNR RTSQNQAARP STNQQTATQP QITTNAIEDT QESNSAEPEI 120
121 SREPSRTNTV EHRTPEPRLS LDDSKEEEMV VHRIPSVTRA AEVARIPDVP IILFMGGPGG 180
181 GKTRHAARVA DSLADNGLVH ICMPDIIRTA LGKYKDKYPE WKEANEHYIR GELIPNQLAL 240
241 TLLKAEMGRH PDAMGFFLEG YPREARQVED FERQVKSVNM ALILDYDERT LREHMERRGL 300
301 GMEIIDQKIK EFKQKTLPSA KYFDDQKLLH LIPGEKDDQV IYEKMKALVV KALETGVPVL 360
361 AALPPMAADR HHIASPAAPE SVIPSPIMAE SELTNSVLDT NVANHETINH DLSTSAIREP 420
421 RSVAQTPLHS SGSGAGGGGG DVESKPATPA VQTPDESRAT STARSRESAR KEKLATPDSK 480
481 NAADTRNKTS SNGSSEIVSA AELGEPVGLP NNAPVILVLG APGSQKNDIS RRIAQKYDGF 540
541 TMLSMGDILR KKINNEKNDE MWDKVSKKMN NGDPIPTKMC RTVLYEELHS RGTSNWGYVI 600
601 EGYPKSPDQL VDLEHSLQRT DLAILIDCTE QFCLEVINKR NRENKRSDDD SDAVRSRMEY 660
661 FKKNTLPMLK TLDDKGKLRV VDGDADPDTV FKEVVQVIDR TLYVEDDGDG TSLGDSKKGT 720
721 LNSSLNSST |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
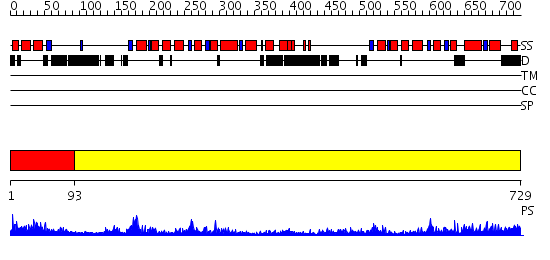
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..92] | 1.364989 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [93..729] | 71.522879 | Crystal Structure Analysis of ClpB |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)