













| General Information: |
|
| Name(s) found: |
ptp-1 /
CE31450
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 589 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoskeleton
[IEA cytoplasm [IEA extrinsic to membrane [IEA |
| Biological Process: |
protein amino acid dephosphorylation
[IEA dephosphorylation [IEA |
| Molecular Function: |
protein binding
[IEA protein tyrosine phosphatase activity [IEA phosphatase activity [IEA cytoskeletal protein binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRRRRTARTS ESSAPSLRQQ RLSKEAIYYG TQESCDEKSW TPSMACTSTS PGIHASTASV 60
61 RPVSSGSTPN GASRKSANSG YSGYGYATQT QQPTSTTNAS YSPYLNGTIS RSSGAAVAKA 120
121 AARGLPPTNQ QAYNTSSPRN SVASYSSFAS AGIGGSPPRS KRSPQSNKSS SPVGEDQVVT 180
181 IKMRPDRHGR FGFNVKGGAD QNYPVIVSRV APGSSADKCQ PRLNEGDQVL FIDGRDVSTM 240
241 SHDHVVQFIR SARSGLNGGE LHLTIRPNVY RLGEEVDEPD STMVPEPARV ADSVPNSDKL 300
301 SKSLQLLADS LNSGKVVDHF EMLYRKKPGM SMNICRLTAN LAKNRYRDVC PYDDTRVTLQ 360
361 ASPSGDYINA NYVNMEIPSS GIVNRYIACQ GPLAHTSSDF WVMVWEQHCT TIVMLTTITE 420
421 RGRVKCHQYW PRVFETQEYG RLMIKCIKDK QTTNCCYREF SIRDRNSSEE RRVTQMQYIA 480
481 WPDHGVPDDP KHFIQFVDEV RKARQGSVDP IVVHCSAGIG RTGVLILMET AACLVESNEP 540
541 VYPLDIVRTM RDQRAMLIQT PGQYTFVCES ILRAYHDGTI KPLAEYSKR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
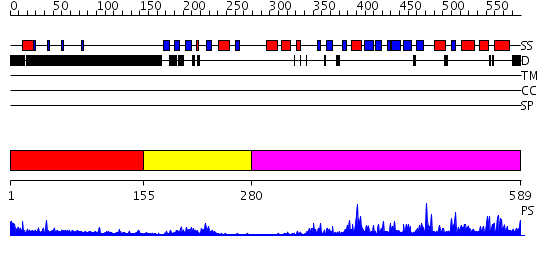
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..154] | 7.173997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [155..279] | 11.154902 | Solution structure of PDZ domain of Protein tyrosine phosphatase, non-receptor type 4 | |
| 3 | View Details | [280..589] | 75.045757 | No description for 2i75A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)