













| General Information: |
|
| Name(s) found: |
lin-1 /
CE31440
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 441 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
negative regulation of vulval development
[IMP regulation of cell fate specification [IMP vulval development [IGI hermaphrodite genitalia development [IMP embryonic development ending in birth or egg hatching [IMP regulation of transcription, DNA-dependent [IEA |
| Molecular Function: |
transcription factor activity
[IEA sequence-specific DNA binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNHIDLLKVK KEPPSSSEEA EEEESPKHTI EGILDIRKKE MNVSDLMCTR SSTPPTSSSV 60
61 DSIITLWQFL LELLQQDQNG DIIEWTRGTD GEFRLIDAEA VARKWGQRKA KPHMNYDKLS 120
121 RALRYYYEKN IIKKVIGKKF VYRFVTTDAH APPTADFSSN MNMKMCYVKD EKDIRHEIPS 180
181 FMTSLQAPPP PPPPPQNPRG NTDFSALSLL GTDSPTTHSV STPSPTDSVC SPSSSVASSA 240
241 TPSTSSPVDE SRQCRKRSLS PSTTSSTTAP PPPPQPPTKK GMKPNPLNLT ATSNFSLQPS 300
301 ISSPLLMLQQ HHQNSPLFQA QISQLYTYAA LASAGLYGPQ ISPHLASQSP FRSPLVTPKN 360
361 LGLGELGSSG RTPGLGESQV FQFPPVSAFQ ATNPLLNTFS NLISPMAPFM MPPSQSSTSF 420
421 KFPSSTDSLK TPTVPIKMPT L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
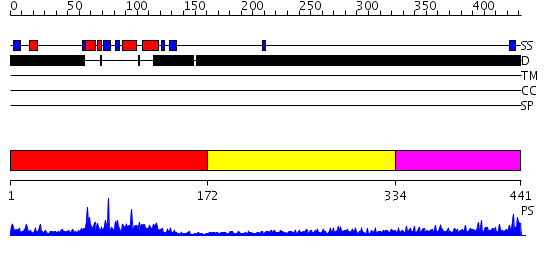
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | 43.221849 | ETS-1 transcription factor, residues 331-440 | |
| 2 | View Details | [172..333] | 1.05 | No description for 2j63A was found. | |
| 3 | View Details | [334..441] | 5.167996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)