













| General Information: |
|
| Name(s) found: |
elt-2 /
CE31430
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 433 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
regulation of epithelial cell differentiation
[IMP locomotory behavior [IMP nematode larval development [IMP endoderm development [IMP digestion [IMP regulation of transcription, DNA-dependent [IEA positive regulation of transcription from RNA polymerase II promoter [IDA growth [IMP reproduction [IMP |
| Molecular Function: |
transcription factor activity
[IEA zinc ion binding [IEA RNA polymerase II transcription factor activity, enhancer binding [IDA double-stranded DNA binding [IDA sequence-specific DNA binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDNNYNDNVN GWAEMEPSQP MGGLRLPTQN MDPPEQNNES QLSELPRMKI DNDYASPIER 60
61 QSVITSGTNN YEPKVETVTS FFHTGIDYSN FGMLDQTTMQ PFYPLYSGIP VNTLGTFSGY 120
121 TNSIYDKPSL YDPSIPTINI PSTYPTVAPT YECVKCSQSC GAGMKAVNGG MMCVNCSTPK 180
181 TTYSPPVAYS TSLGQPPILE IPSEQPTAKI AKQSSKKSSS SNRGSNGSAS RRQGLVCSNC 240
241 NGTNTTLWRR NAEGDPVCNA CGLYFKLHHI PRPTSMKKEG ALQTRKRKSK SGDSSTPSTS 300
301 RARERKFERA SSSTEKAQRS SNRRAGSAKA DRELSTAAVA AATATYVSHA DLYPVSSAAV 360
361 TLPDQTYSNY YQWNTAATAG LMMVPNDQNY VYAATNYQTG LRPADNIQVH VMPVQDDETK 420
421 AAARDLEAVD GDS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
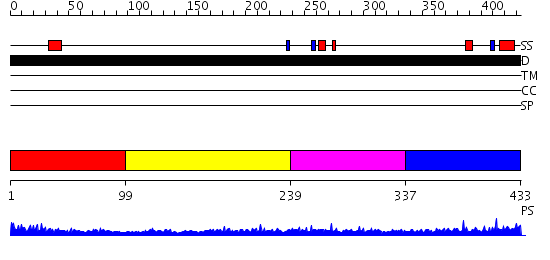
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..98] | 2.166996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [99..238] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [239..336] | 9.30103 | Erythroid transcription factor GATA-1 | |
| 4 | View Details | [337..433] | 4.171991 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)