













| General Information: |
|
| Name(s) found: |
div-1 /
CE28840
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 581 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
DNA replication
[IEA embryonic development ending in birth or egg hatching [IMP |
| Molecular Function: |
DNA binding
[IEA DNA-directed DNA polymerase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAYEFDLEDF NDNLEPFGFN CPAKDHFIEK LEGLCKTFRK SGEDIVDDVV SVMSNLGQKS 60
61 VDSGILAKLE ENLEAQAQKT VHTPRSVKKP ARGRVPLAEK SGFILSGDPD DIEKPSHHLT 120
121 NSAIAKLEGH FLDFSPFPGS PANEKFLKRA DPSHVVTTIR GKKYQDKGLK GSTKSDDTIR 180
181 IISKTPSTLY GGDKSSMVIE AKAQRMADIA HRIQKSFEEI VDWGNPSIPS VDVVYTYGQV 240
241 IHDETKDNEK FGEHSVALMI NDEDGTMIRM DFSKMTEDIT LFPGQIIAVR GTNETGEELQ 300
301 VDKIFQPAAL PVNPVETDTT KEIWFACGPY TATDNCGYEH LCELLDKVVA EKPDILMLAG 360
361 PFVDKKNTFL NKPTFNITYD NLLEDLLLKV KETLVGTKTQ VIIQPNASRD LCVPPVFPSA 420
421 PFQQNRKLDK IKKELIFVAD PCIFRISGVE VAMTSSEPIQ ALSNTEFHRS ANQENIDRVA 480
481 RLSSHLLTQQ CMYPLEPTEV PASMGDLLDV CCIGSTPHIV FAPTKLAPSA KCVNGSVFIN 540
541 SSTLAKGPTG NYAKMSINLN AGELMPGETV ADYAQIQILK I |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
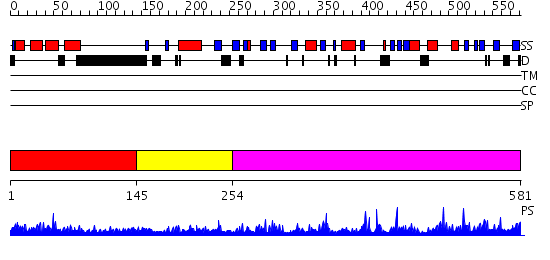
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..144] | 1.062985 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [145..253] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [254..581] | 1.7 | Structure of Vacuolar Protein Sorting 29 from Cryptosporidium Parvum |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)