













| General Information: |
|
| Name(s) found: |
pde-4 /
CE28641
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 626 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
signal transduction
[IEA |
| Molecular Function: |
catalytic activity
[IEA 3',5'-cyclic-nucleotide phosphodiesterase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCGSRRHLRR NEDGMNGVAR RKQFKLRPWQ STALPSRHDH YSCAGSATAA DGLGGAHLTP 60
61 SLSSSVHARR ESFLYRASDD LREASSLRPV SRASSIASNE HGHGDDLIVT PFAQLLASLR 120
121 NVRSNLISIT NIQNSDDSRH ANRSAKRPPL HNIELPDDVV HCAHDTLEEL DWCLDQLETI 180
181 QTHRSVSEMA SSKFRKMLNK ELSHFAESSK SGTQVSKFLI TTYMDKEEDE PSIEIEVPTE 240
241 VQGPSTSGPM TLSILKKAQT AAMNKISGVR KLRAPSHDGH VPEYGVNCAR EIAVHMQRLD 300
301 DWGPDVFKID ELSKNHSLTV VTFSLLRQRN LFKTFEIHQS TLVTYLLNLE HHYRNNHYHN 360
361 FIHAADVAQS MHVLLMSPVL TEVFTDLEVL AAIFAGAVHD VDHPGFTNQY LINSNNELAI 420
421 MYNDESVLEQ HHLAVAFKLL QDSNCDFLAN LSRKQRLQFR KIVIDMVLAT DMSKHMSLLA 480
481 DLKTMVEAKK VAGNNVIVLD KYNDKIQVLQ SMIHLADLSN PTKPIELYQQ WNQRIMEEYW 540
541 RQGDKEKELG LEISPMCDRG NVTIEKSQVG FIDYIVHPLY ETWADLVYPD AQNILDQLEE 600
601 NREWYQSRIP EEPDTARTVT EDDEHK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
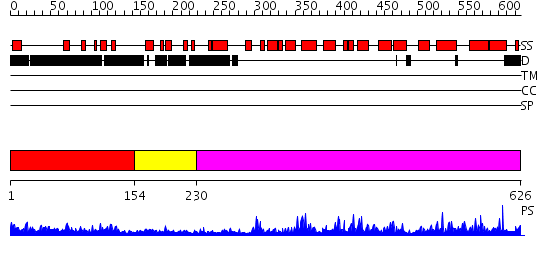
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..153] | 5.061987 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [154..229] | 2.13499 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [230..626] | 125.0 | Catalytic domain of cyclic nucleotide phosphodiesterase pde4d |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)