













| General Information: |
|
| Name(s) found: |
tdc-1 /
CE28344
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 705 amino acids |
Gene Ontology: |
|
| Cellular Component: |
neuronal cell body
[IDA axon [IDA |
| Biological Process: |
carboxylic acid metabolic process
[IEA |
| Molecular Function: |
pyridoxal phosphate binding
[IEA carboxy-lyase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVYGLGEALK NLNSYCQERT TRIRNSLSPS RPSMSEATAT GSSSSSRAST TIPSTPNMDV 60
61 TPTVEDPRQN DNNASGMTRD EFRQYGKETV DYIVDYLENI QKRRVVPAIE PGYLKDLIPS 120
121 EAPNTPESFE SVMEDFEKLI MPGITHWQHP RFHAYFPAGN SFPSIIADML SDAIGCVGFS 180
181 WAACPAMTEL ELIMLDWFGK MIGLPAEFLP LTENGKGGGV IQSSASECNF VTLLAARFEV 240
241 MKELRQRFPF VEEGLLLSKL IAYCSKEAHS SVEKACMIGM VKLRILETDS KFRLRGDTLR 300
301 NAIQEDRNLG LIPFFVSTTL GTTSCCSFDV LSEIGPICKE NELWLHVDAA YSGSAFICPE 360
361 FRPLMNGIEY AMSFNTNPNK WLLINFDCST MWVRDRFKLT QALVVDPLYL QHSWMDKSID 420
421 YRHWGIPLSR RFRSLKLWFV IRMYGIDGLQ KYIREHVRLA KKMETLLRAD AKFEIVNEVI 480
481 MGLVCFRMKG DDELNQTLLT RLNASGRIHM VPASLGDRFV IRFCVCAENA TDKDIEVAYE 540
541 IIAQATQHVL HDSVKAVIAE EDEEAVALEE MVADLNITET PEKCLTRQNS ANAAESGQKL 600
601 ERQLSKEEIL AQKQHESLAK KRSFLVRMVS DPKCYNPKIV RHLNMANHRK MSQDLYRDRT 660
661 LMQTISHSQR PNRLSQSPGS AGSAFFDDDD DRIVADVQTG LQTPI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
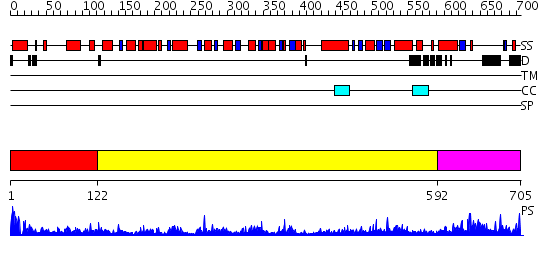
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..121] | 109.0 | DOPA decarboxylase | |
| 2 | View Details | [122..591] | 21.0 | Serine hydroxymethyltransferase | |
| 3 | View Details | [592..705] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)