













| General Information: |
|
| Name(s) found: |
nhr-91 /
CE27779
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 474 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
positive regulation of vulval development
[IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP regulation of transcription, DNA-dependent [IEA |
| Molecular Function: |
DNA binding
[IEA transcription factor activity [IEA zinc ion binding [IEA steroid hormone receptor activity [IEA sequence-specific DNA binding [IEA steroid binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDVADSTSET PPVFDHDNLA LAAQTNLLQF YLTHLASTLV PPMKLDMRDS MTPSFSQTES 60
61 PNSETDDSTS AANKVLFSIL GTPTTDSTSN AVSKSSSKLC SVCGDKSTGL HYGAATCEGC 120
121 KGFFKRSVQN KKVYHCSQDN CCEIDKQNRN RCQSCRFRKC ISKGMLTEAV REDRMPGGRN 180
181 GNSIYSNYKQ RRSIIRKTKD YVEEQERRPF QSLPSGKKLI KELVEMDCLD RLINLKGLRI 240
241 NPSANCDIAP ACKRLTRIGD EIVEQLVEWT KTLPFFDELP VEAHTHLLTQ RWAELVLLSA 300
301 GYYACSVFTP DSPDTTEMID ETDEISFTNP QVNLRLLQNR LSLVLGKEIP LEHVAKEAGP 360
361 LVTRFTTLLH SFSNLKVSPE AYVCIKAITL LHLSADSTLD RSIIDKVNTL QDHFVKTLQI 420
421 HLHQPGEDAQ TTSLAQILDW LPDLRNASSV LLHSKMFYVP FLLCKNPRRL VFDE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
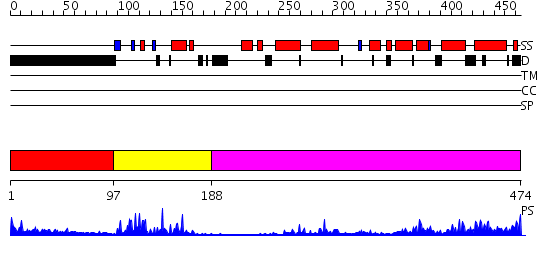
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..96] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [97..187] | 21.39794 | Human Liver Receptor Homologue DNA-Binding Domain (hLRH-1 DBD) in Complex with dsDNA from the hCYP7A1 Promoter | |
| 3 | View Details | [188..474] | 36.30103 | Crystal Structure of the RARbeta/RXRalpha Ligand Binding Domain Heterodimer in Complex with 9-cis Retinoic Acid and a Fragment of the TRAP220 Coactivator |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)