













| General Information: |
|
| Name(s) found: |
CE23855
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 518 amino acids |
Gene Ontology: |
|
| Cellular Component: |
HslUV protease complex
[IEA cytoplasm [IEA |
| Biological Process: |
protein transport
[IEA |
| Molecular Function: |
nucleotide binding
[IEA ATPase activity [IEA ATP binding [IEA protein binding [IEA nucleoside-triphosphatase activity [IEA HslUV protease activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFRIASSSYS QFIASANPSI QTRCISIARI DQKIKRGNSD RRVIHAKKFT KQTDSPASVA 60
61 GDGFILSKTF LQNAIAHQPP SGLTPVGRDG GRDEKKLILH PKEIVEHLNK YVVGQEEAKK 120
121 YLAVAVYQHY RRVENNLRVT EQWMLSEAVA AAKERKKMRK QNPEEEEYYP EYVQKSQRQI 180
181 LKDLEKRQDM ILDKSNMILL GASGTGKTFM TQKLAEVLDV PIVICDCTTL TQAGYVGDDV 240
241 DTVIQKLLAE AMGDIEKCQR GIVFLDEFDK IYTSSDPLHT SGNRDVSGKG VQQALLKLVE 300
301 GSLVKVRDPL APNSKVTIDT SNILFIASGA FSNIEHIVAR RMDKRSLGFL SATSPHKLGD 360
361 QDTTEKLRDS DEEIVSKARN EMIKQCDQGD LISFGMIPEL VGRFPVIVPF HCLDKTHLMS 420
421 VLTEPRGSLI AQTKKFFENE NVELRFSPAA IEAIAEMAVK RKTGARALKS IVEKAVMNAK 480
481 YEVPGSDVKC VEITDQNLKD HSFTVIENQN VIGTENTN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
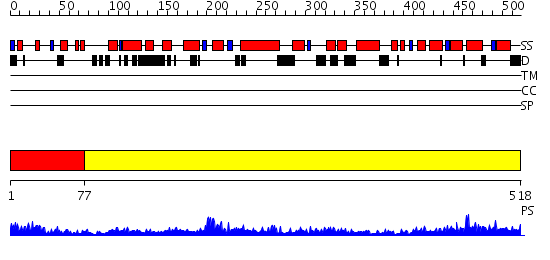
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..76] | 2.457994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [77..518] | 69.0 | Crystal structure of helicobacter pylori ClpX |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.77 |
Source: Reynolds et al. (2008)