













| General Information: |
|
| Name(s) found: |
ptp-1 /
CE23603
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 624 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoskeleton
[IEA cytoplasm [IEA extrinsic to membrane [IEA |
| Biological Process: |
protein amino acid dephosphorylation
[IEA dephosphorylation [IEA |
| Molecular Function: |
protein binding
[IEA protein tyrosine phosphatase activity [IEA phosphatase activity [IEA cytoskeletal protein binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEVPAFVGKA CLKLKGLCVN VWTATVATCQ ENAEIDEKLP PRAFRESSAP SLRQQRLSKE 60
61 AIYYGTQESC DEKSWTPSMA CTSTSPGIHA STASVRPVSS GSTPNGASRK SANSGYSGYG 120
121 YATQTQQPTS TTNASYSPYL NGTISRSSGA AVAKAAARGL PPTNQQAYNT SSPRNSVASY 180
181 SSFASAGIGG SPPRSKRSPQ SNKSSSPVGE DQVVTIKMRP DRHGRFGFNV KGGADQNYPV 240
241 IVSRVAPGSS ADKCQPRLNE GDQVLFIDGR DVSTMSHDHV VQFIRSARSG LNGGELHLTI 300
301 RPNVYRLGEE VDEPDSTMVP EPARVADSVP NSDKLSKSLQ LLADSLNSGK VVDHFEMLYR 360
361 KKPGMSMNIC RLTANLAKNR YRDVCPYDDT RVTLQASPSG DYINANYVNM EIPSSGIVNR 420
421 YIACQGPLAH TSSDFWVMVW EQHCTTIVML TTITERGRVK CHQYWPRVFE TQEYGRLMIK 480
481 CIKDKQTTNC CYREFSIRDR NSSEERRVTQ MQYIAWPDHG VPDDPKHFIQ FVDEVRKARQ 540
541 GSVDPIVVHC SAGIGRTGVL ILMETAACLV ESNEPVYPLD IVRTMRDQRA MLIQTPGQYT 600
601 FVCESILRAY HDGTIKPLAE YSKR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
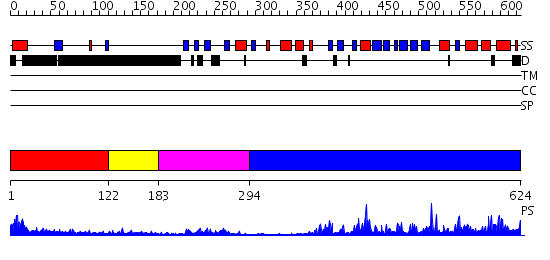
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..121] | 4.221849 | Solution Structure of The forth PDZ Domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 Protein) | |
| 2 | View Details | [122..182] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [183..293] | 3.39794 | Solution structure of the fourth PDZ domain of KIAA1095 protein | |
| 4 | View Details | [294..624] | 76.522879 | No description for 2i75A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)