













| General Information: |
|
| Name(s) found: |
abce-1 /
CE21713
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 610 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Gram-negative-bacterium-type cell wall
[IEA membrane [IEA outer membrane-bounded periplasmic space [IEA |
| Biological Process: |
embryonic development ending in birth or egg hatching
[IMP positive regulation of growth rate [IMP gamete generation [IMP transport [IEA nodulation [IEA growth [IMP positive regulation of multicellular organism growth [IMP cell division [IEA cobalt ion transport [IEA electron transport [IEA locomotory behavior [IMP nematode larval development [IMP nitrate transport [IEA phosphonate transport [IEA polyamine transport [IEA cytochrome complex assembly [IEA molybdate ion transport [IEA |
| Molecular Function: |
ATPase activity
[IEA nucleotide binding [IEA ATP binding [IEA ATPase activity, coupled to transmembrane movement of substances [IEA nitrate transmembrane transporter activity [IEA electron carrier activity [IEA molybdate ion transmembrane transporter activity [IEA nucleoside-triphosphatase activity [IEA polyamine-transporting ATPase activity [IEA iron-sulfur cluster binding [IEA phosphonate transmembrane-transporting ATPase activity [IEA transporter activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRKGPLIKN NETDVPLRIA IVEKDRCKPK NCGLACKRAC PVNRQGKQCI VVEATSTISQ 60
61 ISEILCIGCG ICVKKCPYDA IKIINLPANL ANETTHRYSQ NSFKLHRLPT PRCGEVLGLV 120
121 GTNGIGKSTA LKILAGKQKP NLGNFQKEQE WTTIINHFRG SELQNYFTRI LEDTLKCVIK 180
181 PQYVDQIPRA AKGTVEKNLT RKHDNDNLNS VIDQMELRGL LDREIDQLSG GELQRFAIAM 240
241 CCVQKADVYM FDEPSSYLDV KQRLKAAAII RERVSDTNYV VVVEHDLAVL DYLSDFICCL 300
301 YGVPGVYGVV TLPSGVREGI NMFLEGFIRT ENMRFRESKL SFKTSEQQED IKRTGNIKYP 360
361 SMSKTLGNFH LDVEAGDFSD SEIIVMLGEN GTGKTTMIKM MAGSLKPEDE NTELPHVSIS 420
421 YKPQKISPKS ETTVRFMLHD KIQNMYEHPQ FKTDVMNPLM MEQLLDRNVK ELSGGELQRV 480
481 ALALCLGKTA SLYLIDEPSA YLDSEQRLHA AKVIKRFIMH AKKTAFVVEH DFIMATYLAD 540
541 RVVVFEGQPS VKCTACKPQS LLEGMNRFLK MLDITFRRDQ ETYRPRINKL DSVKDVDQKK 600
601 SGQFFFLDDN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
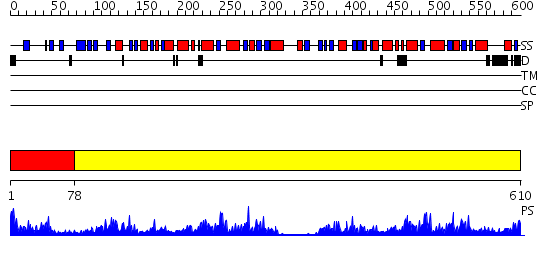
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..77] | 2.42 | Crystal structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus | |
| 2 | View Details | [78..610] | 97.39794 | RNase-L Inhibitor |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)