













| General Information: |
|
| Name(s) found: |
unc-62 /
CE21219
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 564 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
oocyte construction
[IMP locomotory behavior [IMP hermaphrodite genitalia development [IMP embryonic development ending in birth or egg hatching [IMP regulation of transcription, DNA-dependent [IEA oviposition [IMP cell migration [IMP positive regulation of multicellular organism growth [IMP |
| Molecular Function: |
transcription factor activity
[IEA sequence-specific DNA binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGDSKVWAH ASADTWAVQN GISTYDLDTS SIKREKRDHN EQFNDGYGPP PGSASADPAS 60
61 YIADPAAFYN LYTNMGGAPT STPMMHHEMG EAMKRDKESI YAHPLYPLLV LLFEKCELAT 120
121 STPRDTSRDG STSSDVCSSA SFKDDLNEFV RHTQENADKQ YYVPNPQLDQ IMLQSIQMLR 180
181 FHLLELEKVH ELCDNFCNRY VVCLKGKMPL DIVGDERASS SQPPMSPGSM GHLGHSSSPS 240
241 MAGGATPMHY PPPYEPQSVP LPENAGVMGG HPMEGSSMAY SMAGMAAAAA SSSSSSNQAG 300
301 DHPLANGGTL HSTAGASQTL LPIAVSSPST CSSGGLRQDS TPLSGETPMG HANGNSMDSI 360
361 SEAGDEFSVC GSNDDGRDSV LSDSANGSQN GKRKVPKVFS KEAITKFRAW LFHNLTHPYP 420
421 SEEQKKQLAK ETGLTILQVN NWFINARRRI VQPMIDQNNR AGRSGQMNVC KNRRRNRSEQ 480
481 SPGPSPDSGS DSGANYSPDP SSLAASTAMP YPAEFYMQRT MPYGGFPSFT NPAMPFMNPM 540
541 MGFQVAPTVD ALSQQWVDLS APHE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
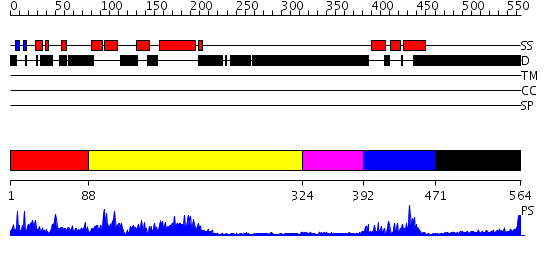
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [88..323] | 4.52597 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [324..391] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [392..470] | 11.69897 | Solution structure of the homeobox domain of human homeobox protein PKNOX1 | |
| 5 | View Details | [471..564] | 3.165995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)