













| General Information: |
|
| Name(s) found: |
mrg-1 /
CE20213
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 335 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA autosome [IDA chromatin [IEA |
| Biological Process: |
germ cell development
[IMP negative regulation of transcription from RNA polymerase II promoter [IMP hermaphrodite genitalia development [IMP embryonic development ending in birth or egg hatching [IMP post-embryonic body morphogenesis [IMP chromatin assembly or disassembly [IEA gamete generation [IMP positive regulation of transcription from RNA polymerase II promoter [IGI reproduction [IMP |
| Molecular Function: |
chromatin binding
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSKKNFEVG ENVACIYKGK PYDAKITDIK TNSDGKELYC VHFKGWNNRY DEKIPVGEEK 60
61 DRIFKGTASE YAEKHNAELP TTALKPKKKS LAAEAPRDDR DDTPGTSKGK KAKSVTIAPV 120
121 MTADDMKVEL PKPLRKILID DYDLVCRYFI NIVPHEYSVD QIIEDYIKTI PVSNEQMRTV 180
181 DDLLIEYEEA DIKITNLALI CTARGLVDYF NVTLGYQLLY KFERPQYNDL VKKRAMEKGI 240
241 DITNPTALQD SGFRPSQEYG IVHFLRMLAK LPDYLKLTQW NDHVINRIMI GVHDLIVFLN 300
301 KNHGKYYRGS SDYQGASNDY YRRSLAADDG VGANQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
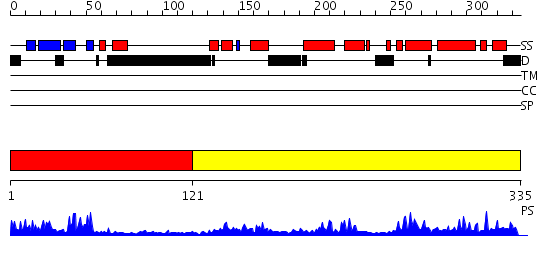
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..120] | 18.522879 | Solution Structure of the Tudor Domain from Mouse Hypothetical Protein Homologous to Histone Acetyltransferase | |
| 2 | View Details | [121..335] | 50.69897 | Crystal Structure of the MRG15 MRG domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)