













| General Information: |
|
| Name(s) found: |
CE18278
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 591 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
protein amino acid dephosphorylation
[IEA dephosphorylation [IEA |
| Molecular Function: |
protein tyrosine phosphatase activity
[IEA phosphatase activity [IEA cytoskeletal protein binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQRRKGTRGN SQDDQQASRR LAKKPSDEKV DEKDKKKGGV LKRLGFLLHN EKKEEVGSTQ 60
61 KRRKPKRGTE RKRACSHPVW NPDKKIHFNN EKTHAIIPLK GDPLSDVQKK VFFKFASDAV 120
121 KKNPPEYSVE FMTKVKPYPG QPMERKIFDA NPTKNRYKDV VCNDITRVIL SDGGDGDYIH 180
181 ANYVNGLNAP FILTQGATAA TVIDFWRMVV HTKTAYIVML CEVMEDGKAK CAQYYPEKAG 240
241 EAMTFGAWTI LCSLEDDKDA NIIKRTLSVR NVDNGKEHIL KHLHTKSWPD RCVPNSTMAL 300
301 LRMLYIVRTA SGPVTVHCSA GIGRTGTFVA IEACLQILTD GKELDLLGTC RALRNSRAGS 360
361 IQVDIQYMAL VQILINYGKD NGYWDDSDLD DRVELLTWNI QQFIQTRGKV DHVPTTPSTP 420
421 ALNIIPADAK PVPVVPKEAT PHHSPAEKAQ ECMEKICEKI LKPIKKDKEC KEHKSPKTEH 480
481 PKECKDHPLP LETRSSSEDG PDMEDQKTQA TQKVEIKEET AAQNDNISMI SLPNEPLLSI 540
541 PVVRKNSKDG NKDSCDTKES KETKGHEKKN SKEGNFKLVK KQSANRSQYF L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
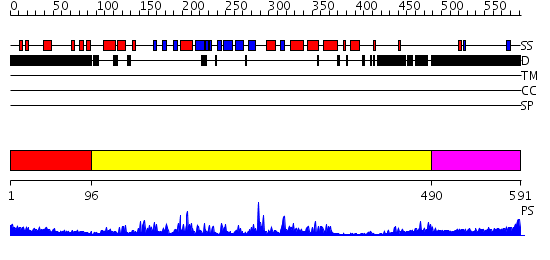
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..95] | 71.0 | Tyrosine phosphatase; Tyrosine phoshatase shp-2 | |
| 2 | View Details | [96..489] | 107.0 | Crystal structure of the phosphatase domains of human PTP SIGMA | |
| 3 | View Details | [490..591] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)