













| General Information: |
|
| Name(s) found: |
CE15651
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 337 amino acids |
Gene Ontology: |
|
| Cellular Component: |
pyruvate dehydrogenase complex
[IEA oxoglutarate dehydrogenase complex [IEA |
| Biological Process: |
metabolic process
[IEA glycolysis [IEA tricarboxylic acid cycle [IEA pyruvate metabolic process [IEA |
| Molecular Function: |
dihydrolipoyllysine-residue succinyltransferase activity
[IEA protein binding [IEA acyltransferase activity [IEA dihydrolipoyllysine-residue acetyltransferase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLAGSQALRH LSTAAQNQGA CGPAVKLLLI QYGLENRKID GTGPKNKNIL KGDVMKIVEA 60
61 EKLKPVAHHA HAPKETHIEN KSIEKKSDIF GANNRSLRHH QDIPLSNIRA TIAKRLTASK 120
121 QQIPHEYQGV DVRIDDILAL RQKLKKSGTA VSLNDFIIKA AALALRSVPT VNVRWTPEGI 180
181 GLGSVDISVA VATPTGLITP IVENSDILGV LAISSKVKEL SGLARESKLK PQQFQGGSFT 240
241 ISNLGMFGSV TNFTAIINPP QCAILTIGGT RSEVVSVDGQ LETQKLMGVN LCFDGRAISE 300
301 ECAKRFLLHF SESLSDPELL IAEPLSPELD FDFSRLL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
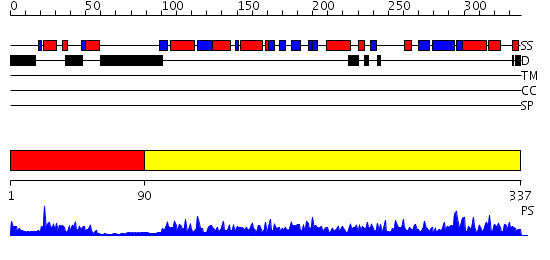
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..89] | 9.522879 | Solution structure of the e3_binding domain of dihydrolipoamide branched chaintransacylase | |
| 2 | View Details | [90..337] | 84.522879 | Dihydrolipoamide succinyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)