













| General Information: |
|
| Name(s) found: |
lin-11 /
CE15150
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 405 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
positive regulation of vulval development
[IMP regulation of cell differentiation [IMP positive regulation of growth rate [IMP oviposition [IMP regulation of transcription, DNA-dependent [IEA cell fate specification [IMP regulation of cell migration [IMP axonal fasciculation [IMP |
| Molecular Function: |
transcription factor activity
[IEA zinc ion binding [IEA sequence-specific DNA binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHSSSSFIIT SLEEEEKKPP AHHLHQQSIE DVGSVTSSAT LLLLDSATWM MPSSTTQPHI 60
61 SEISGNECAA CAQPILDRYV FTVLGKCWHQ SCLRCCDCRA PMSMTCFSRD GLILCKTDFS 120
121 RRYSQRCAGC DGKLEKEDLV RRARDKVFHI RCFQCSVCQR LLDTGDQLYI MEGNRFVCQS 180
181 DFQTATKTST PTSIHRPVSN GSECNSDVEE DNVDACDEVG LDDGEGDCGK DNSDDSNSAK 240
241 RRGPRTTIKA KQLETLKNAF AATPKPTRHI REQLAAETGL NMRVIQVWFQ NRRSKERRMK 300
301 QLRFGGYRQS RRPRRDDIVD MFPNDQQFYP PPPPSNVQFF CDPYTTSPNN PETIQMAPQF 360
361 AVPTENMNMV PEPYTEQSAT PPEFNEDTFA CIYSTDLGKP TPVSW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
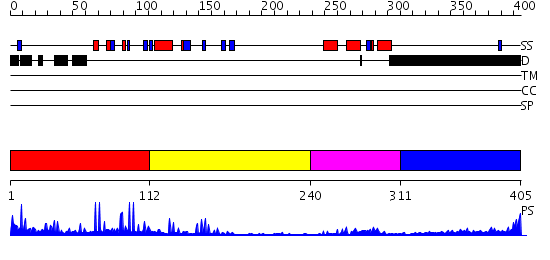
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..111] | 6.0 | Cysteine-rich (intestinal) protein, CRP, CRIP | |
| 2 | View Details | [112..239] | 19.39794 | No description for 2dfyX was found. | |
| 3 | View Details | [240..310] | 6.39794 | DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF THE ANTENNAPEDIA HOMEODOMAIN FROM DROSOPHILA IN SOLUTION BY 1H NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | |
| 4 | View Details | [311..405] | 4.235997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)