













| General Information: |
|
| Name(s) found: |
sur-5 /
CE04713
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 700 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
propionate catabolic process, 2-methylcitrate cycle
[IEA metabolic process [IEA menaquinone biosynthetic process [IEA |
| Molecular Function: |
acetate-CoA ligase activity
[IEA AMP binding [IEA catalytic activity [IEA propionate-CoA ligase activity [IEA o-succinylbenzoate-CoA ligase activity [IEA ligase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTAVSANGKT TEKHENGAHT NGTTNGTTNG SMNGNEISHV QKLQPVYYKP PQNLETFELS 60
61 LRNHFEEKTN KKFADYREFH RFTCDNYGIF WEDLLKLSDV KLHQNYNQVI DHNLKINERP 120
121 RWFNGATLNY TENVIERGTA TDIAVLNASI EETVTEYTYD NLRKDVYRIA TSLRNYGIGP 180
181 GDTVCGFVPN TYDTLVAVFA TAAVGAAWCS ASVDFGPAGV LDRFRQVHPK VLFTVNHVTY 240
241 KKKLIDQTDK INEIVKELPT LEKIVVSDTF TSVKFDATKY NQSDKFSSLE EFKTPIADVV 300
301 LPFVYTPVPF SDPLFVMFSS GTTGIPKAMV HTVGGTLLKH IEEHLVQGDS KKHDRMFFYT 360
361 TCGWMMYNWM ISFLYSKGSV VLFDECPLAP DTHIIMKIAA KTQSTMIGMG AKLYDEYLRL 420
421 QIPFNTLYDL SKIHTVYSTG SPLKKECFAY INTYIAPGAL IASISGGTDI IGCFVGGIKS 480
481 LSITPGECQC LFLGMDIKSF NYMDEEIINS DEQGELVCVT PFPSMPSHFL NDTDGKKYRD 540
541 AYFARLEPFW AHGDFVRVNH STGGVEMLGR SDATLNRGGV RIGTAEIYSV VEKIPHIADC 600
601 IVAGRLVEEG MDEEVLLFVK MVPGQELTHS IQAAIVSKLR NDMSPRHVPN KIYAVDDIPY 660
661 TSSGKKVEVA VKQIVSGKAV QKASSIRNPE SLDHFVQYRL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
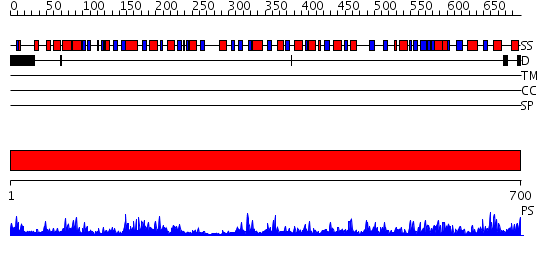
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..700] | 171.0 | No description for 2p2bA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)