













| General Information: |
|
| Name(s) found: |
CE02370
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 309 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
estrogen biosynthetic process
[IEA poly-hydroxybutyrate biosynthetic process [IEA metabolic process [IEA fatty acid biosynthetic process [IEA |
| Molecular Function: |
estradiol 17-beta-dehydrogenase activity
[IEA 3-oxoacyl-[acyl-carrier-protein] reductase activity [IEA oxidoreductase activity [IEA NAD or NADH binding [IEA 3-hydroxybutyrate dehydrogenase activity [IEA acetoacetyl-CoA reductase activity [IEA 2-deoxy-D-gluconate 3-dehydrogenase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MACKNPKKFF PIRKSPVLRD GAFKGKLVLV TGGGTGIGKA IATTFAHLRA TVVIAARRME 60
61 KLEQTARDIT KITGGTCEPF QMDIKDPGMV SDAFDKIDMK FGKVPEILVN NAAGNFIMAT 120
121 ELLSSNAYGT IIDIVLKGTF NVTTELGKRC IQNKTGASIT SITAGYARAG APFIVPSAVS 180
181 KAGVETMTKS LATEWSKYGL RFNAVSPGPI PTKGAWGRLN SGEMGDIAEK MKFLNPEGRV 240
241 GSPEEVANLV AFISSDHMSF LNGAIIDLDG GQQHFNHGSH MGDFLHSWDH KNWEDAENLI 300
301 RGRTGKEKA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
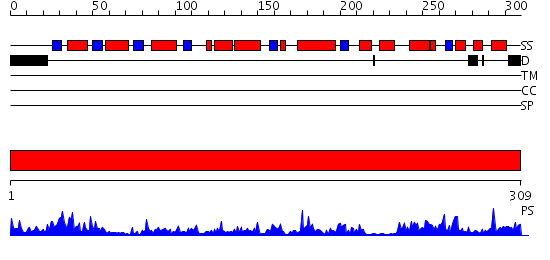
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..309] | 56.0 | Structure of human DECR ternary complex |
Functions predicted (by domain):
| # | Gene Ontology predictions |
| 1 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.70 |
Source: Reynolds et al. (2008)