













| General Information: |
|
| Name(s) found: |
cct-1 /
CE02319
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 549 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
hermaphrodite genitalia development
[IMP nematode larval development [IMP embryonic development ending in birth or egg hatching [IMP positive regulation of growth rate [IMP cellular protein metabolic process [IEA protein folding [IEA reproduction [IMP growth [IMP |
| Molecular Function: |
ATP binding
[IEA protein binding [IEA unfolded protein binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASAGDSILA LTGKRTTGQG IRSQNVTAAV AIANIVKSSL GPVGLDKMLV DDVGDVIVTN 60
61 DGATILKQLE VEHPAGKVLV ELAQLQDEEV GDGTTSVVIV AAELLKRADE LVKQKVHPTT 120
121 IINGYRLACK EAVKYISENI SFTSDSIGRQ SVVNAAKTSM SSKIIGPDAD FFGELVVDAA 180
181 EAVRVENNGK VTYPINAVNV LKAHGKSARE SVLVKGYALN CTVASQAMPL RVQNAKIACL 240
241 DFSLMKAKMH LGISVVVEDP AKLEAIRREE FDITKRRIDK ILKAGANVVL TTGGIDDLCL 300
301 KQFVESGAMA VRRCKKSDLK RIAKATGATL TVSLATLEGD EAFDASLLGH ADEIVQERIS 360
361 DDELILIKGP KSRTASSIIL RGANDVMLDE MERSVHDSLC VVRRVLESKK LVAGGGAVET 420
421 SLSLFLETYA QTLSSREQLA VAEFASALLI IPKVLASNAA RDSTDLVTKL RAYHSKAQLI 480
481 PQLQHLKWAG LDLEEGTIRD NKEAGILEPA LSKVKSLKFA TEAAITILRI DDLIKLDKQE 540
541 PLGGDDCHA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
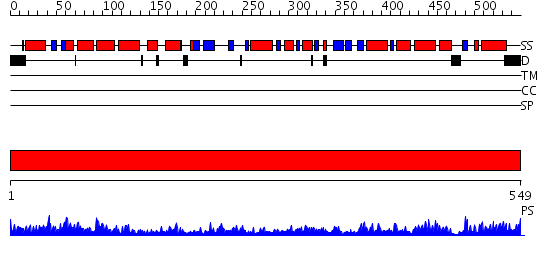
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..549] | 138.0 | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form of single mutant) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)